3THJ
 
 | |
1FHU
 
 | |
1F8M
 
 | | CRYSTAL STRUCTURE OF 3-BROMOPYRUVATE MODIFIED ISOCITRATE LYASE (ICL) FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ISOCITRATE LYASE, MAGNESIUM ION, PYRUVIC ACID | | Authors: | Sharma, V, Sharma, S, Hoener zu Bentrup, K, McKinney, J.D, Russell, D.G, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-06-30 | | Release date: | 2000-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1FH1
 
 | |
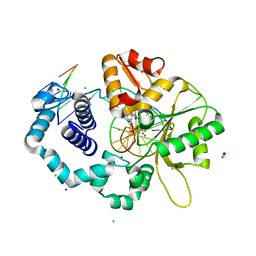
7K96
 
 | |
1FQW
 
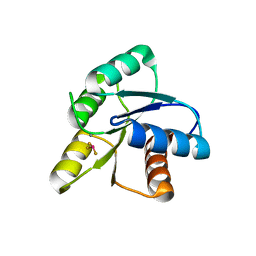 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
1HHK
 
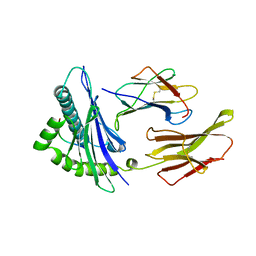 | | THE ANTIGENIC IDENTITY OF PEPTIDE(SLASH)MHC COMPLEXES: A COMPARISON OF THE CONFORMATION OF FIVE PEPTIDES PRESENTED BY HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), NONAMERIC PEPTIDE FROM HTLV-1 TAX PROTEIN (RESIDUES 11-19) | | Authors: | Madden, D.R, Garboczi, D.N, Wiley, D.C. | | Deposit date: | 1993-06-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2.
Cell(Cambridge,Mass.), 75, 1993
|
|
3V76
 
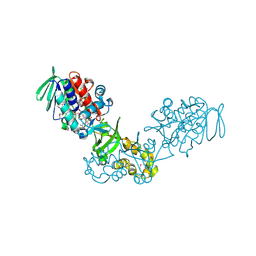 | | The crystal structure of a flavoprotein from Sinorhizobium meliloti | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavoprotein | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The crystal structure of a flavoprotein from Sinorhizobium meliloti
TO BE PUBLISHED
|
|
1ET4
 
 | |
2RN9
 
 | | Solution structure of human apoCox17 | | Descriptor: | Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
2RNB
 
 | | Solution structure of human Cu(I)Cox17 | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
1J6W
 
 | | CRYSTAL STRUCTURE OF HAEMOPHILUS INFLUENZAE LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
3U4F
 
 | | Crystal structure of a mandelate racemase (muconate lactonizing enzyme family protein) from Roseovarius nubinhibens | | Descriptor: | GUANIDINE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Eswaramoorthy, S, Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a mandelate racemase (muconate lactonizing enzyme family protein) from Roseovarius nubinhibens
To be Published, 2011
|
|
3U5T
 
 | | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Sinorhizobium meliloti | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Sinorhizobium meliloti
To be Published
|
|
3U4J
 
 | | Crystal structure of NAD-dependent aldehyde dehydrogenase from Sinorhizobium meliloti | | Descriptor: | CALCIUM ION, NAD-dependent aldehyde dehydrogenase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-08 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NAD-dependent aldehyde dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3U9D
 
 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-24) of drosophila Ciboulot and the C-terminal domain (residues 13-44) of bovine Thymosin-beta4, bound to G-actin-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly.
Embo J., 31, 2012
|
|
3UP8
 
 | | Crystal structure of a putative 2,5-diketo-D-gluconic acid reductase B | | Descriptor: | ACETATE ION, Putative 2,5-diketo-D-gluconic acid reductase B | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a putative 2,5-diketo-D-gluconic acid reductase B
To be Published
|
|
3V2H
 
 | | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti | | Descriptor: | D-beta-hydroxybutyrate dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3V4C
 
 | | Crystal structure of a semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Aldehyde dehydrogenase (NADP+) | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of a semialdehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
1EYF
 
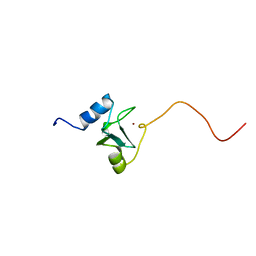 | | REFINED STRUCTURE OF THE DNA METHYL PHOSPHOTRIESTER REPAIR DOMAIN OF E. COLI ADA | | Descriptor: | ADA REGULATORY PROTEIN, ZINC ION | | Authors: | Lin, Y, Dotsch, V, Wintner, T, Peariso, K, Myers, L.C, Penner-Hahn, J.E, Verdine, G.L, Wagner, G. | | Deposit date: | 2000-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the functional switch of the E. coli Ada protein
Biochemistry, 40, 2001
|
|
1ILP
 
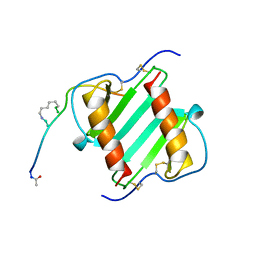 | |
1IRS
 
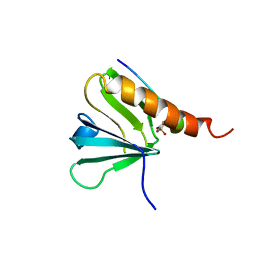 | | IRS-1 PTB DOMAIN COMPLEXED WITH A IL-4 RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | IL-4 RECEPTOR PHOSPHOPEPTIDE, IRS-1 | | Authors: | Zhou, M.-M, Huang, B, Olejniczak, E.T, Meadows, R.P, Shuker, S.B, Miyazaki, M, Trub, T, Shoelson, S.E, Feisk, S.W. | | Deposit date: | 1996-03-22 | | Release date: | 1997-05-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain.
Nat.Struct.Biol., 3, 1996
|
|
7KPY
 
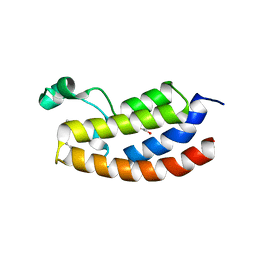 | | Crystal structure of CBP bromodomain liganded with UMB298 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloranyl-4-methoxy-phenyl)ethyl]-~{N}-cyclohexyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)imidazo[1,2-a]pyridin-3-amine, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Dimethylisoxazole-Attached Imidazo[1,2- a ]pyridines as Potent and Selective CBP/P300 Inhibitors.
J.Med.Chem., 64, 2021
|
|
3V2G
 
 | | Crystal structure of a dehydrogenase/reductase from Sinorhizobium meliloti 1021 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, SULFATE ION | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-01-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a dehydrogenase/reductase from Sinorhizobium meliloti 1021
To be Published
|
|
3UOE
 
 | | The crystal structure of dehydrogenase from Sinorhizobium meliloti | | Descriptor: | Dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-16 | | Release date: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The crystal structure of dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
