3LD8
 
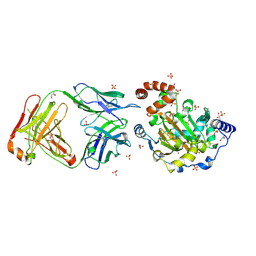 | | Structure of JMJD6 and Fab Fragments | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, GLYCEROL, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5K5B
 
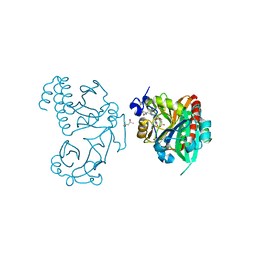 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans BphP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, ... | | Authors: | Takala, H, Edlund, P, Claesson, E, Ihalainen, J.A, Westenhoff, S. | | Deposit date: | 2016-05-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
4XYH
 
 | |
6OPC
 
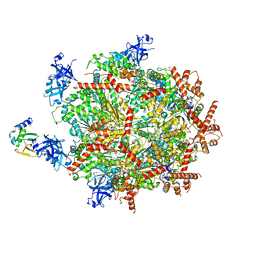 | | Cdc48 Hexamer in a complex with substrate and Shp1(Ubx Domain) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Cooney, I, Han, H, Stewart, M, Carson, R.H, Hansen, D, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Cdc48 segregase in the act of unfolding an authentic substrate.
Science, 365, 2019
|
|
468D
 
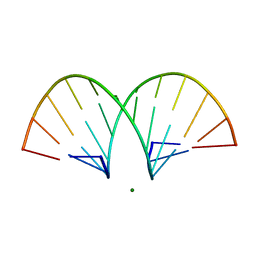 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
4Y34
 
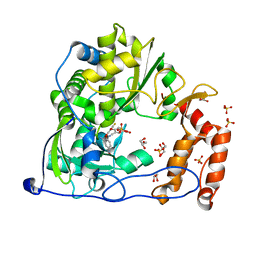 | | Crystal Structure of Coxsackievirus B3 3D polymerase in complex with GPC-N143 | | Descriptor: | 2,2'-[(4-fluorobenzene-1,2-diyl)bis(oxy)]bis(5-nitrobenzonitrile), 3D polymerase, GLYCEROL, ... | | Authors: | Vives-Adrian, L, Ferrer-Orta, C, Cerdaguer, N. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
3P8X
 
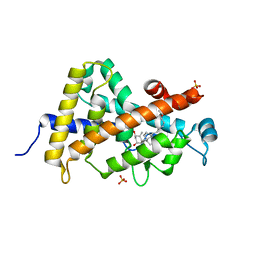 | | Synthesis, Structure, and Biological Activity of des-Side Chain Analogues of 1alpha,25-Dihydroxyvitamin D3 with Substituents at C-18 | | Descriptor: | (1R,3S,5Z)-5-{(2E)-2-[(3aR,7aS)-7a-(7-hydroxy-7-methyloctyl)octahydro-4H-inden-4-ylidene]ethylidene}-4-methylidenecyclohexane-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Rochel, N, Sato, Y, Moras, D. | | Deposit date: | 2010-10-15 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis, Structure, and Biological Activity of des-Side Chain Analogues of 1 ,25-Dihydroxyvitamin D3 with Substituents at C-18
Chemmedchem, 6, 2011
|
|
4Y3C
 
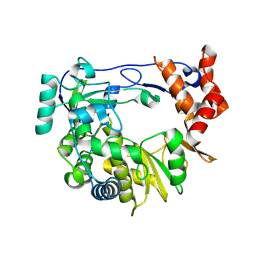 | | I304V 3D polymerase mutant of EMCV | | Descriptor: | 3D polymerase, CHLORIDE ION, GLYCEROL | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
3OZ0
 
 | | PPAR Delta in complex with azppard02 | | Descriptor: | Peroxisome proliferator-activated receptor delta, [4-({(1S)-1-[(2,4-dichlorophenyl)carbamoyl]-1,3-dihydro-2H-isoindol-2-yl}methyl)-2-methylphenoxy]acetic acid | | Authors: | Ogg, D. | | Deposit date: | 2010-09-24 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of isoindoline and tetrahydroisoquinoline derivatives as potent, selective PPARδ agonists
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4Y2C
 
 | | M300V 3D polymerase mutant of EMCV | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
3LDB
 
 | | Structure of JMJD6 complexd with ALPHA-KETOGLUTARATE and Fab Fragment. | | Descriptor: | 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XUF
 
 | | CRYSTAL STRUCTURE OF MACHE-Y337A-TZ2PA6 ANTI COMPLEX (1 MTH) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-6-PHENYL-5-[6-[1-[2-[(1,2,3,4-TETRAHYDRO-9-ACRIDINYL)AMINO]ETHYL]-1H-1,2,3-TRIAZOL-4-YL]HEXYL]-PHENANTHRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Radic, Z, Taylor, P, Marchot, P. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conformational Remodeling of Femtomolar Inhibitor-Acetylcholinesterase Complexes in the Crystalline State
J.Am.Chem.Soc., 132, 2010
|
|
6P59
 
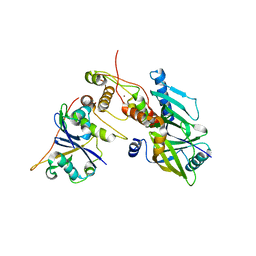 | | Crystal structure of SIVrcm Vif-CBFbeta-ELOB-ELOC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Core-binding factor subunit beta, Elongin-B, ... | | Authors: | Binning, J.M, Chesarino, N.M, Emerman, M, Gross, J.D. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.942214 Å) | | Cite: | Structural Basis for a Species-Specific Determinant of an SIV Vif Protein toward Hominid APOBEC3G Antagonism.
Cell Host Microbe, 26, 2019
|
|
6P6G
 
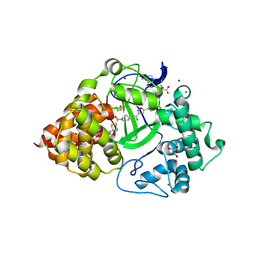 | | Co-crystal Structure of human SMYD3 with Isoxazole Amides Inhibitors | | Descriptor: | 5-cyclopropyl-N-{1-[({trans-4-[(4,4,4-trifluorobutyl)amino]cyclohexyl}methyl)sulfonyl]piperidin-4-yl}-1,2-oxazole-3-carboxamide, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Wang, L. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Isoxazole Amides as Potent and Selective SMYD3 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
4A76
 
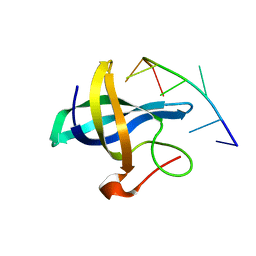 | | The Lin28b Cold shock domain in complex with heptathymidine | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP)-3', LIN28 COLD SHOCK DOMAIN | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2011-11-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
6P6K
 
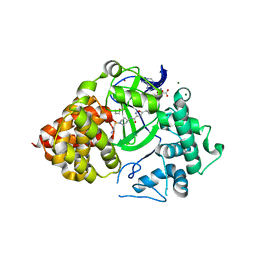 | |
4YH0
 
 | |
2RE8
 
 | |
4YDA
 
 | |
4YDB
 
 | |
6P74
 
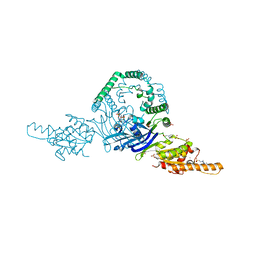 | | OLD nuclease from Thermus Scotoductus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PLATINUM (II) ION, Putative ATP-dependent endonuclease of the OLD family, ... | | Authors: | Chappie, J.S, Schiltz, C.J. | | Deposit date: | 2019-06-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The full-length structure of Thermus scotoductus OLD defines the ATP hydrolysis properties and catalytic mechanism of Class 1 OLD family nucleases.
Nucleic Acids Res., 48, 2020
|
|
4YFQ
 
 | |
7R1F
 
 | |
7R0E
 
 | |
7R42
 
 | | Bovine complex I in the presence of IM1761092, active class ii (Composite map) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-carbamimidoyl-3-[2-(3-chloranyl-4-iodanyl-phenyl)ethyl]guanidine, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
