4FJZ
 
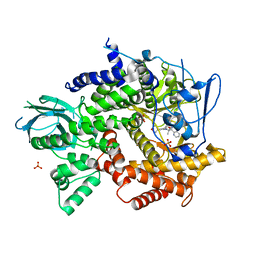 | | Crystal structure of PI3K-gamma in complex with pyrrolo-pyridine inhibitor 63 | | Descriptor: | 1'-[7-fluoro-3-methyl-2-(pyridin-2-yl)quinolin-4-yl]-6'-(morpholin-4-yl)-1',2,2',3,5,6-hexahydrospiro[pyran-4,3'-pyrrolo[3,2-b]pyridine], Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and in Vivo Evaluation of Dual PI3K-beta/delta inhibitors
J.Med.Chem., 55, 2012
|
|
5QOW
 
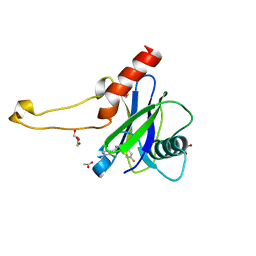 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z2895259675 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N-[(4-fluorophenyl)methyl]-1-methyl-1H-pyrazole-4-carboxamide, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3NLB
 
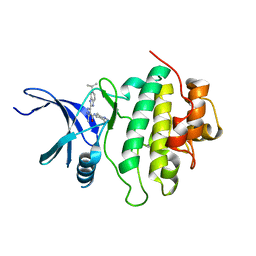 | | Novel kinase profile highlights the temporal basis of context dependent checkpoint pathways to cell death | | Descriptor: | 3-methyl-5-[5-(1-methylethyl)-1H-benzimidazol-2-yl]-N-(1-methylpiperidin-4-yl)-1H-pyrazole-4-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Massey, A.J, Borgognoni, J, Bentley, C, Foloppe, N, Fiumana, A, Walmsley, L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent cell cycle checkpoint abrogation by a novel kinase inhibitor
Plos One, 5, 2010
|
|
3AIS
 
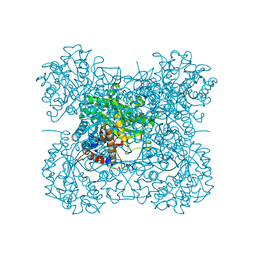 | | Crystal structure of a mutant beta-glucosidase in wheat complexed with DIMBOA-Glc | | Descriptor: | (2S)-2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one, Beta-glucosidase, beta-D-glucopyranose | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
4OWN
 
 | | Anthranilate phosphoribosyl transferase from Mycobacterium tuberculosis in complex with 5-fluoroanthranilate, PRPP and Magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-amino-5-fluorobenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Cookson, T.V.M, Short, F.L, Lott, J.S. | | Deposit date: | 2014-02-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.
Biochem.J., 461, 2014
|
|
3CR6
 
 | |
5QOZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with PB1787571279 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FIM
 
 | | Cryo-EM structure of the tirzepatide (LY3298176)-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,human glucagon like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-07-31 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
2V0K
 
 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
7Y1R
 
 | | Human L-TGF-beta1 in complex with the anchor protein LRRC33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
2E5P
 
 | | Solution structure of the TUDOR domain of PHD finger protein 1 (PHF1 protein) | | Descriptor: | PHD finger protein 1 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TUDOR domain of PHD finger protein 1 (PHF1 protein)
To be Published
|
|
3VP4
 
 | | Crystal structure of human glutaminase in complex with inhibitor 4 | | Descriptor: | 5,5'-butane-1,4-diylbis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5QJ5
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with Z44592329 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1GR7
 
 | | Crystal structure of the double mutant Cys3Ser/Ser100Pro from Pseudomonas Aeruginosa at 1.8 A resolution | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Okvist, M, Bonander, N, Sandberg, A, Karlsson, B.G, Krengel, U, Xue, Y, Sjolin, L. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the double azurin mutant Cys3Ser/Ser100Pro from Pseudomonas aeruginosa at 1.8 A resolution: its folding-unfolding energy and unfolding kinetics.
Biochim.Biophys.Acta, 1596, 2002
|
|
3RN6
 
 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with zinc and isoguanine | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, 6-amino-3,7-dihydro-2H-purin-2-one, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Rescue of the orphan enzyme isoguanine deaminase.
Biochemistry, 50, 2011
|
|
5QK3
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z239136710 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QOQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z2895259680 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethyl-N-[(4-fluorophenyl)methyl]-1,2,4-oxadiazole-5-carboxamide, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3VP3
 
 | | Crystal structure of human glutaminase in complex with inhibitor 3 | | Descriptor: | 5,5'-pentane-1,5-diylbis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5AWB
 
 | | Crystal structure of human TLR8 in complex with N1-3-aminomethylbenzyl (meta-amine) | | Descriptor: | 1-[[3-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Human TLR8-Specific Agonists with Augmented Potency and Adjuvanticity.
J.Med.Chem., 58, 2015
|
|
5AXH
 
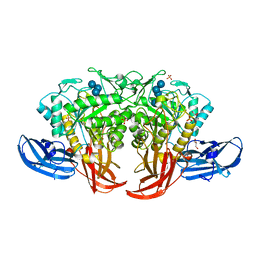 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose | | Descriptor: | Dextranase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
1VPM
 
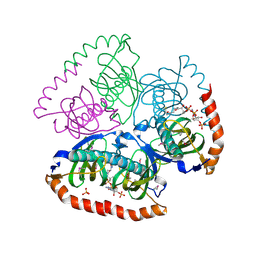 | |
3VT9
 
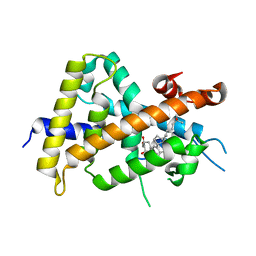 | | Crystal structures of rat VDR-LBD with W282R mutation | | Descriptor: | (1R,2Z,3R,5E,7E,9beta,17beta)-2-(2-hydroxyethylidene)-17-[(2R)-6-hydroxy-6-methylheptan-2-yl]-9-(prop-2-en-1-yl)-9,10-secoestra-5,7-diene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
5QK0
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1270312110 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,5-dimethyl-1H-pyrazol-4-yl)aniline, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
3AIQ
 
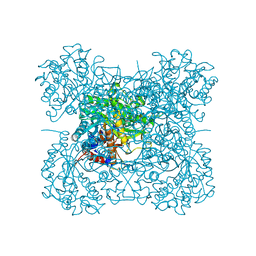 | | Crystal structure of beta-glucosidase in wheat complexed with an aglycone DIMBOA | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, Beta-glucosidase | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
1R7A
 
 | | Sucrose Phosphorylase from Bifidobacterium adolescentis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, sucrose phosphorylase | | Authors: | Sprogoe, D, van den Broek, L.A.M, Mirza, O, Kastrup, J.S, Voragen, A.G.J, Gajhede, M, Skov, L.K. | | Deposit date: | 2003-10-21 | | Release date: | 2004-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis.
Biochemistry, 43, 2004
|
|
