7KW0
 
 | | Non-ribosomal didomain (stabilised glycine-PCP-C) acceptor bound state | | Descriptor: | N-{2-[(2-aminoethyl)sulfanyl]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
7L15
 
 | | Marsupial T cell receptor Spl_118 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, T cell receptor gamma chain, ... | | Authors: | Wegrecki, M, Kannan Sivaraman, K, Praveena, T, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The molecular assembly of the marsupial gamma mu T cell receptor defines a third T cell lineage.
Science, 371, 2021
|
|
7KVW
 
 | | Non-ribosomal didomain (holo-PCP-C) acceptor bound state | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
7KW2
 
 | | Non-ribosomal didomain (holo-PCP-C) acceptor bound state, R2577G | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
7KQ7
 
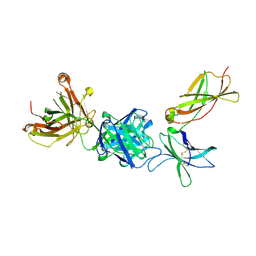 | | Crystal structure of IL21R in complex with an antibody Fab fragment | | Descriptor: | Antibody heavy chain, Antibody light chain, Interleukin-21 receptor | | Authors: | Mosyak, L, Svenson, K. | | Deposit date: | 2020-11-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Combining random mutagenesis, structure-guided design and next-generation sequencing to mitigate polyreactivity of an anti-IL-21R antibody.
Mabs, 13, 2021
|
|
7KYL
 
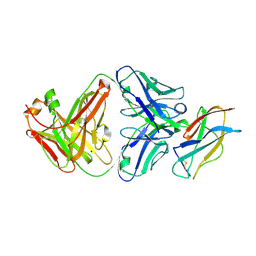 | | Powassan virus Envelope protein DIII in complex with neutralizing Fab POWV-80 | | Descriptor: | CHLORIDE ION, Envelope protein domain III, POWV-80 Fab heavy chain, ... | | Authors: | Errico, J.M, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broadly neutralizing monoclonal antibodies protect against multiple tick-borne flaviviruses.
J.Exp.Med., 218, 2021
|
|
7KWU
 
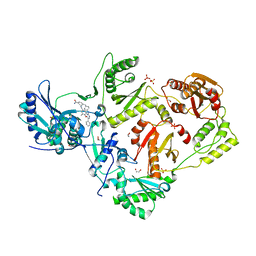 | | Crystal Structure of HIV-1 RT in Complex with 16c (K07-15) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-5-(pyridin-4-yl)pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzamide, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Arnold, E. | | Deposit date: | 2020-12-02 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 2,4,5-Trisubstituted Pyrimidines as Potent HIV-1 NNRTIs: Rational Design, Synthesis, Activity Evaluation, and Crystallographic Studies.
J.Med.Chem., 64, 2021
|
|
7KJZ
 
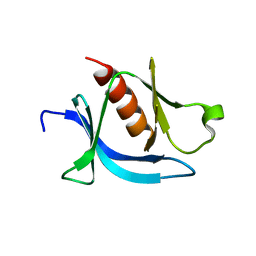 | | crystal structure of PLEKHA7 PH domain biding inositol-tetraphosphate | | Descriptor: | 1,2-ETHANEDIOL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-26 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
7KWW
 
 | | X-ray Crystal Structure of PlyCB Mutant K59H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KNG
 
 | | 2.10A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-2 Y7F) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, DTY-ASP-TYR-PRO-GLY-ASP-PHE-CYS-TYR-LEU-TYR-GLY-THR-CYS, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7KL5
 
 | | Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues: Phe4246 to Val4271 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-1, ... | | Authors: | Fisher, A.J, Ames, J.B, Yu, Q. | | Deposit date: | 2020-10-29 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues Phe4246-Val4271 Reveals a Fifth Calcium Binding Site.
Biochemistry, 60, 2021
|
|
7KK7
 
 | | crystal structure of ligand-free PLEKHA7 PH domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
7KJO
 
 | |
7KWT
 
 | | X-ray Crystal Structure of PlyCB Mutant Y28H | | Descriptor: | PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KNF
 
 | | 1.80A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-1 NHOH) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, DTY-ASP-TYR-PRO-GLY-ASP-HIS-CYS-TYR-LEU-TYR-GLY-THR, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7KWY
 
 | | X-ray Crystal Structure of PlyCB Mutant R66K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KIB
 
 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 4 | | Descriptor: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-amino-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Palte, R.L, Hayes, R.P. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7L5I
 
 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 7.0 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
7KGX
 
 | | Structure of PQS Response Protein PqsE in Complex with 4-(3-(2-methyl-2-morpholinobutyl)ureido)-N-(thiazol-2-yl)benzamide | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-({[(2R)-2-methyl-2-(morpholin-4-yl)butyl]carbamoyl}amino)-N-(1,3-thiazol-2-yl)benzamide, FE (III) ION | | Authors: | Jeffrey, P.D, Taylor, I.R, Bassler, B.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor Mimetic Mutations in the Pseudomonas aeruginosa PqsE Enzyme Reveal a Protein-Protein Interaction with the Quorum-Sensing Receptor RhlR That Is Vital for Virulence Factor Production.
Acs Chem.Biol., 16, 2021
|
|
7KP1
 
 | | CD1a-42:2 SM binary complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, T-cell surface glycoprotein CD1a, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
7KH5
 
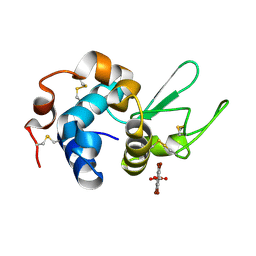 | | Hen Egg White Lysozyme in complex with tetrabromoterephthalic acid | | Descriptor: | 2,3,5,6-tetrabromobenzene-1,4-dicarboxylic acid, Lysozyme C | | Authors: | Truong, J, Nguyen, S. | | Deposit date: | 2020-10-20 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.295 Å) | | Cite: | Simplified heavy-atom derivatization of protein structures via co-crystallization with the MAD tetragon tetrabromoterephthalic acid.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7KP0
 
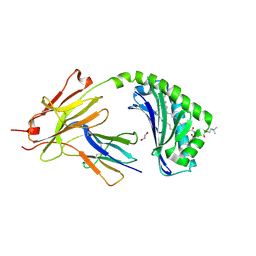 | | CD1a-42:1 SM binary complex | | Descriptor: | (4R,7S)-4-hydroxy-7-[(1S,2E)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-4,9-dioxo-3,5-dioxa-8-aza-4lambda~5~-phosphadotriacontan-1-aminium, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
7KZI
 
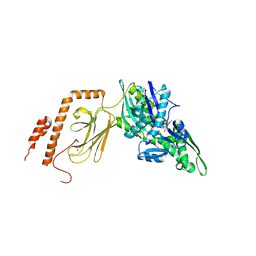 | | Intermediate state (QQQ) of near full-length DnaK alternatively fused with a substrate peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chaperone protein DnaK fused with substrate peptide,Chaperone protein DnaK fused with substrate peptide, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Intermediates in allosteric equilibria of DnaK-ATP interactions with substrate peptides
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7KGW
 
 | |
7KOZ
 
 | | CD1a-36:2 SM binary complex | | Descriptor: | (4S,7S,17Z)-4-hydroxy-7-[(1S,2E)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-4,9-dioxo-3,5-dioxa-8-aza-4lambda~5~-phosphahexacos-17-en-1-aminium, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
