1VJV
 
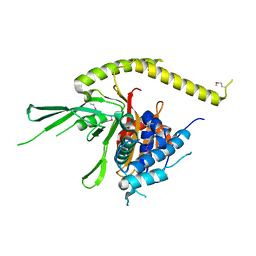 | |
3W9Y
 
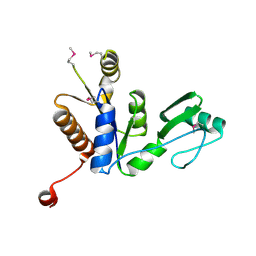 | | Crystal structure of the human DLG1 guanylate kinase domain | | Descriptor: | Disks large homolog 1 | | Authors: | Mori, S, Tezuka, Y, Arakawa, A, Handa, N, Shirouzu, M, Akiyama, T, Yokoyama, S. | | Deposit date: | 2013-04-18 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the guanylate kinase domain from discs large homolog 1 (DLG1/SAP97)
Biochem.Biophys.Res.Commun., 435, 2013
|
|
4ARX
 
 | | Lepidoptera-specific toxin Cry1Ac from Bacillus thuringiensis ssp. kurstaki HD-73 | | Descriptor: | 1,3-DIAMINOPROPANE, GLYCEROL, PESTICIDAL CRYSTAL PROTEIN CRY1AC | | Authors: | Derbyshire, D.J, Carroll, J, Ellar, D.J, Li, J. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Galnac-Dependent Receptor Recognition by B. Thuringiensis Toxin Cry1Ac
To be Published
|
|
7KQS
 
 | | A 1.68-A resolution 3-fluoro-L-tyrosine bound crystal structure of heme-dependent tyrosine hydroxylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
5TKU
 
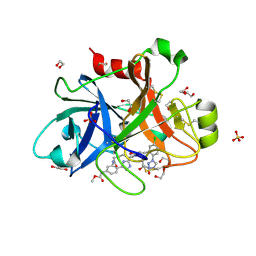 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR METHYL ((15S)-15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2-PROPENOYL)AMINO)-9-OXO-8,17,19-TRIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE | | Descriptor: | 1,2-ETHANEDIOL, Factor XIa (Light Chain), METHYL ((15S)-15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2-PROPENOYL)AMINO)-9-OXO-8,17,19-TRIAZATRICYCLO[14.2.1.0~2,7~]N ONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-07 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Design of Macrocyclic Factor XIa Inhibitors: Discovery of the Macrocyclic Amide Linker.
J. Med. Chem., 60, 2017
|
|
2V0R
 
 | | crystal structure of a hairpin exchange variant (LTx) of the targeting LINE-1 retrotransposon endonuclease | | Descriptor: | LTX, SULFATE ION | | Authors: | Repanas, K, Zingler, N, Layer, L.E, Schumann, G.G, Perrakis, A, Weichenrieder, O. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determinants for DNA Target Structure Selectivity of the Human Line-1 Retrotransposon Endonuclease
Nucleic Acids Res., 35, 2007
|
|
7KQU
 
 | | A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Davis, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQT
 
 | |
2G54
 
 | |
1VBD
 
 | | POLIOVIRUS (TYPE 1, MAHONEY STRAIN) COMPLEXED WITH R78206 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE PROPYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1AMB
 
 | |
3WI2
 
 | | Crystal structure of PDE10A in complex with inhibitor | | Descriptor: | 2-({[1-phenyl-2-(propan-2-yl)-1H-benzimidazol-6-yl]oxy}methyl)quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y. | | Deposit date: | 2013-09-04 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Design and synthesis of novel benzimidazole derivatives as phosphodiesterase 10A inhibitors with reduced CYP1A2 inhibition.
Bioorg.Med.Chem., 21, 2013
|
|
3C2F
 
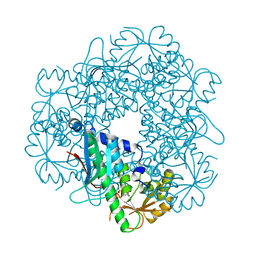 | |
2QHM
 
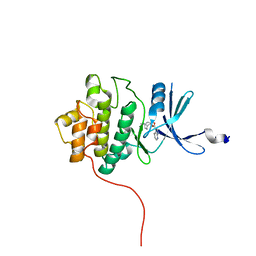 | | crystal structure of Chek1 in complex with inhibitor 2a | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLATE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S. | | Deposit date: | 2007-07-02 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of a pyrazoloquinolinone class of Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4B0G
 
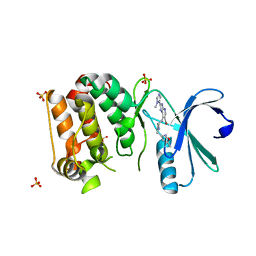 | | Complex of Aurora-A bound to an Imidazopyridine-based inhibitor | | Descriptor: | 6-bromo-2-(1-methyl-1H-imidazol-5-yl)-7-{4-[(5-methyl-1,2-oxazol-3-yl)methyl]piperazin-1-yl}-1H-imidazo[4,5-b]pyridine, AURORA KINASE A, SULFATE ION | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2012-07-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization of Imidazo[4,5-B]Pyridine-Based Kinase Inhibitors: Identification of a Dual Flt3/Aurora Kinase Inhibitor as an Orally Bioavailable Preclinical Development Candidate for the Treatment of Acute Myeloid Leukemia.
J.Med.Chem., 55, 2012
|
|
3OCF
 
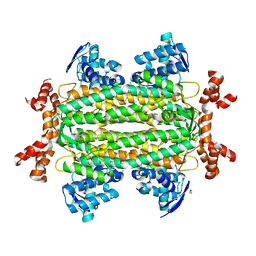 | |
1AFU
 
 | |
1JY2
 
 | | Crystal Structure of the Central Region of Bovine Fibrinogen (E5 fragment) at 1.4 Angstroms Resolution | | Descriptor: | FIBRINOGEN ALPHA CHAIN, FIBRINOGEN BETA CHAIN, FIBRINOGEN GAMMA-B CHAIN | | Authors: | Madrazo, J, Brown, J.H, Litvinovich, S, Dominguez, R, Yakovlev, S, Medved, L, Cohen, C. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the central region of bovine fibrinogen (E5 fragment) at 1.4-A resolution.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5RFL
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102389 | | Descriptor: | 1-acetyl-N-(2-hydroxyphenyl)piperidine-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4BAE
 
 | | Optimisation of pyrroleamides as mycobacterial GyrB ATPase inhibitors: Structure Activity Relationship and in vivo efficacy in the mouse model of tuberculosis | | Descriptor: | 2-[(3S,4R)-4-[(3-bromanyl-4-chloranyl-5-methyl-1H-pyrrol-2-yl)carbonylamino]-3-methoxy-piperidin-1-yl]-4-(2-methyl-1,2,4-triazol-3-yl)-1,3-thiazole-5-carboxylic acid, CALCIUM ION, DNA GYRASE SUBUNIT B, ... | | Authors: | Read, J.A, Gingell, H.G, Madhavapeddi, P. | | Deposit date: | 2012-09-13 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of Pyrrolamides as Mycobacterial Gyrb ATPase Inhibitors: Structure Activity Relationship and in Vivo Efficacy in the Mouse Model of Tuberculosis.
Antimicrob.Agents Chemother., 58, 2014
|
|
7LC4
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with gamma-lactam YU253911 | | Descriptor: | 1-[(2S)-2-{[(2Z)-2-(2-amino-5-chloro-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-3-oxopropyl]-4-{[2-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl]carbamoyl}-2,5-dihydro-1H-pyrazole-3-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2021-01-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A gamma-lactam siderophore antibiotic effective against multidrug-resistant Pseudomonas aeruginosa, Klebsiella pneumoniae, and Acinetobacter spp.
Eur.J.Med.Chem., 220, 2021
|
|
3WVT
 
 | |
4LNB
 
 | | Aspergillus fumigatus protein farnesyltransferase ternary complex with farnesyldiphosphate and ethylenediamine scaffold inhibitor 5 | | Descriptor: | 1,2-ETHANEDIOL, CaaX farnesyltransferase alpha subunit Ram2, CaaX farnesyltransferase beta subunit Ram1, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
3C2A
 
 | | Antibody Fab fragment 447-52D in complex with UG1033 peptide | | Descriptor: | Envelope glycoprotein, Fab 447-52D heavy chain, Fab 447-52D light chain | | Authors: | Dhillon, A.K, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2008-01-24 | | Release date: | 2008-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of an anti-HIV-1 Fab 447-52D-peptide complex from an epitaxially twinned data set
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1A34
 
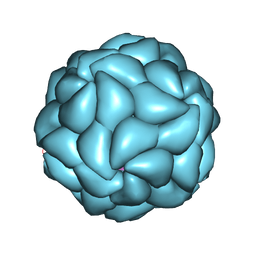 | | SATELLITE TOBACCO MOSAIC VIRUS/RNA COMPLEX | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), SATELLITE TOBACCO MOSAIC VIRUS, ... | | Authors: | Larson, S.B, Day, J, Greenwood, A.J, McPherson, A. | | Deposit date: | 1998-01-28 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Refined structure of satellite tobacco mosaic virus at 1.8 A resolution.
J.Mol.Biol., 277, 1998
|
|
