6CGP
 
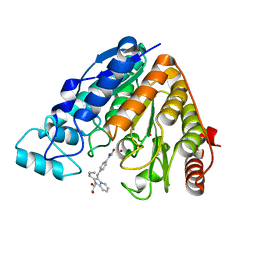 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with MAIP-032 | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-hydroxy-4-[(2-propylimidazo[1,2-a]pyridin-3-yl)amino]benzamide, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2018-02-20 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multicomponent Synthesis and Binding Mode of Imidazo[1,2- a]pyridine-Capped Selective HDAC6 Inhibitors.
Org. Lett., 20, 2018
|
|
8EE3
 
 | |
8Y20
 
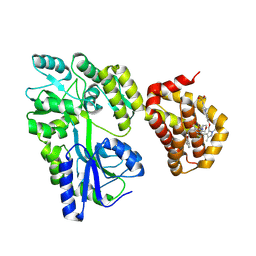 | | Crystal structure of the Mcl-1 in complex with A-1210477 | | Descriptor: | A-1210477, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Deciphering molecular specificity in MCL-1/BAK interaction and its implications for designing potent MCL-1 inhibitors.
Cell Death Differ., 32, 2025
|
|
7L0I
 
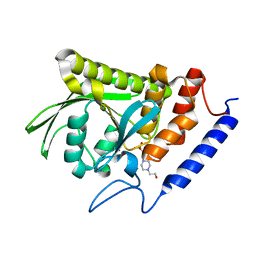 | | Ligand-free YopH G352T | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Protein-tyrosine-phosphatase | | Authors: | Shen, R.D, Hengge, A.C, Johnson, S.J. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Single Residue on the WPD-Loop Affects the pH Dependency of Catalysis in Protein Tyrosine Phosphatases.
Jacs Au, 1, 2021
|
|
5I90
 
 | | Crystal Structure of PvdN from Pseudomonas Aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, PYRIDOXAL-5'-PHOSPHATE, PvdN | | Authors: | Drake, E.J, Gulick, A.M. | | Deposit date: | 2016-02-19 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | 1.2 angstrom resolution crystal structure of the periplasmic aminotransferase PvdN from Pseudomonas aeruginosa.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
6BW2
 
 | | Mcl-1 complexed with small molecules | | Descriptor: | 3-({11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}methyl)benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2017-12-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of Potent and Selective Tricyclic Indole Diazepinone Myeloid Cell Leukemia-1 Inhibitors Using Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6CQ9
 
 | | K2P2.1(TREK-1):ML402 complex | | Descriptor: | CADMIUM ION, HEXADECANE, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2018-03-14 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | K2P2.1 (TREK-1)-activator complexes reveal a cryptic selectivity filter binding site.
Nature, 547, 2017
|
|
5OBK
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-((3,5-dichlorophenyl)sulfonyl)-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Sippel, C, Haehle, A, Bracher, A, Hausch, F. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Chemogenomic Profiling of Human and Microbial FK506-Binding Proteins.
J. Med. Chem., 61, 2018
|
|
5I42
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with a DNA aptamer, AZTTP, and CA(2+) ion | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (38-MER), ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet.
Acs Chem.Biol., 11, 2016
|
|
5O6Y
 
 | | Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide, ... | | Authors: | Fadouloglou, V.E, Kotsifaki, D, Kokkinidis, M. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide
To Be Published
|
|
5O36
 
 | | Japanese encephalitis virus non-structural protein 1' C-terminal domain | | Descriptor: | Japanese encephalitis virus non-structural protein 1' (NS1'),Japanese encephalitis virus non-structural protein 1' (NS1'), N-PROPANOL, SULFATE ION | | Authors: | Thanalai, P, Wright, G.S.A, Antonyuk, S.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Study of the C-Terminal Domain of Nonstructural Protein 1 from Japanese Encephalitis Virus.
J. Virol., 92, 2018
|
|
4CGO
 
 | | Leishmania major N-myristoyltransferase in complex with a thienopyrimidine inhibitor | | Descriptor: | 3-[methyl-[2-[methyl-(1-methylpiperidin-4-yl)amino]thieno[3,2-d]pyrimidin-4-yl]amino]propanenitrile, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
4CGM
 
 | | Leishmania major N-myristoyltransferase in complex with a biphenyl- derivative inhibitor | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, N-[[3-[3-(6,7-dihydro-4H-[1,3]thiazolo[5,4-c]pyridin-5-ylmethyl)phenyl]phenyl]methyl]-2-pyridin-3-yl-ethanamine, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
6CE0
 
 | | Crystal structure of a HIV-1 clade B tier-3 isolate H078.14 UFO-BG Env trimer in complex with broadly neutralizing Fabs PGT124 and 35O22 at 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-05 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (4.602 Å) | | Cite: | HIV-1 vaccine design through minimizing envelope metastability.
Sci Adv, 4, 2018
|
|
4CLX
 
 | |
4CL8
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 2,4-diamino-6-(3-formylphenyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
1YMT
 
 | | Mouse SF-1 LBD | | Descriptor: | 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL, Nuclear receptor 0B2, Steroidogenic factor 1 | | Authors: | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, Juzumiene, D, Bynum, J.M, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Cell(Cambridge,Mass.), 120, 2005
|
|
5IF4
 
 | | Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) inhibitors using Structure-Based Design | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Discovery and biological characterization of potent myeloid cell leukemia-1 inhibitors.
FEBS Lett., 591, 2017
|
|
4XCS
 
 | | Human peroxiredoxin-1 C83S mutant | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, Peroxiredoxin-1 | | Authors: | Cho, K.J, Lee, J.-H, Khan, T.G, Park, Y, Cho, A, Chang, T.-S, Kim, K.H. | | Deposit date: | 2014-12-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Dimeric Human Peroxiredoxin-1 C83S Mutant
Bull.Korean Chem.Soc., 36, 2015
|
|
9EYV
 
 | | Human PRMT5 in complex with AZ compound 12 | | Descriptor: | (1~{S})-~{N}-[(4-fluorophenyl)methyl]-3-oxidanylidene-2-(1~{H}-pyrrolo[3,2-b]pyridin-2-ylmethyl)-1~{H}-isoindole-1-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
7KHT
 
 | | The acyl chains of phosphoinositide PIP3 alter the structure and function of nuclear receptor Steroidogenic Factor-1 (SF-1) | | Descriptor: | (2S)-3-{[(S)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane-1,2-diyl (9E,9'E)di-octadec-9-enoate, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha peptide, Steroidogenic factor 1 | | Authors: | Blind, R.D. | | Deposit date: | 2020-10-22 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | The acyl chains of phosphoinositide PIP3 alter the structure and function of nuclear receptor steroidogenic factor-1.
J.Lipid Res., 62, 2021
|
|
4UUU
 
 | | 1.7 A resolution structure of human cystathionine beta-synthase regulatory domain (del 516-525) in complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, CYSTATHIONINE BETA-SYNTHASE, S-ADENOSYLMETHIONINE | | Authors: | Kopec, J, McCorvie, T.J, Fitzpatrick, F, Strain-Damerell, C, Froese, D.S, Tallant, C, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
J.Biol.Chem., 289, 2014
|
|
8SW5
 
 | | Protein Phosphatase 1 in complex with PP1-specific Phosphatase targeting peptide (PhosTAP) version 1 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PP1-specific Phosphatase-Targeting Peptide version 1, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A protein phosphatase 1 specific phos phatase ta rgeting p eptide (PhosTAP) to identify the PP1 phosphatome.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4CN1
 
 | | GlgE isoform 1 from Streptomyces coelicolor D394A mutant with maltose- 1-phosphate bound | | Descriptor: | ALPHA-1,4-GLUCAN: MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-1-O-phosphono-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
9EZ1
 
 | | Vitamin D receptor in complex with 1,4a,25-trihydroxyvitamin D3 | | Descriptor: | 1,4a,25-trihydroxyvitamin D3, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Hydroxy-1 alpha ,25-Dihydroxyvitamin D 3 : Synthesis and Structure-Function Study.
Biomolecules, 14, 2024
|
|
