5D5M
 
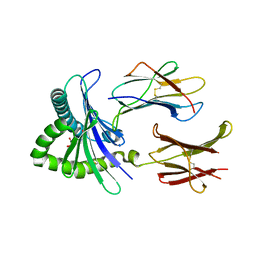 | | Structure of human MR1-5-OP-RU in complex with human MAIT M33.64 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Keller, A.N, Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2015-08-11 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
5D7I
 
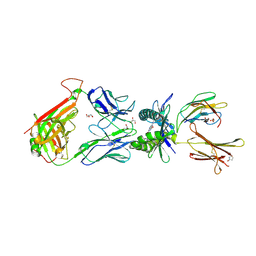 | | Structure of human MR1-Ac-6-FP in complex with human MAIT M33.64 TCR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, M33.64 TCR Alpha Chain, ... | | Authors: | Keller, A.N, Woolley, R.E, Rossjohn, J. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
6V2E
 
 | |
3IVP
 
 | |
3FAY
 
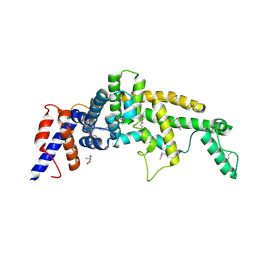 | | Crystal structure of the GAP-related domain of IQGAP1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ras GTPase-activating-like protein IQGAP1 | | Authors: | Kurella, V.B, Richard, J.M, Parke, C.L, Bellamy, H, Worthylake, D.K. | | Deposit date: | 2008-11-18 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the GTPase-activating protein-related domain from IQGAP1.
J.Biol.Chem., 284, 2009
|
|
6YUC
 
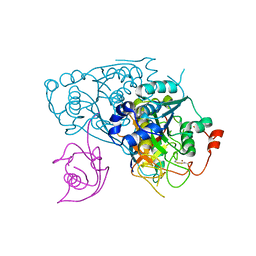 | | Crystal structure of Uba4-Urm1 from Chaetomium thermophilum | | Descriptor: | Adenylyltransferase and sulfurtransferase uba4, Ubiquitin-related modifier 1, ZINC ION | | Authors: | Grudnik, P, Pabis, M, Ethiraju Ravichandran, K, Glatt, S. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Molecular basis for the bifunctional Uba4-Urm1 sulfur-relay system in tRNA thiolation and ubiquitin-like conjugation.
Embo J., 39, 2020
|
|
6Z6S
 
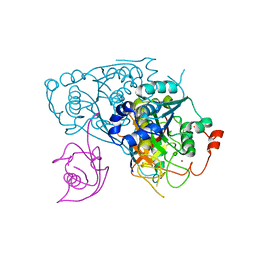 | | Crystal structure of Uba4-Urm1 from Chaetomium thermophilum | | Descriptor: | Adenylyltransferase and sulfurtransferase uba4, Ubiquitin-related modifier 1, ZINC ION | | Authors: | Grudnik, P, Pabis, M, Ethiraju Ravichandran, K, Glatt, S. | | Deposit date: | 2020-05-29 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Molecular basis for the bifunctional Uba4-Urm1 sulfur-relay system in tRNA thiolation and ubiquitin-like conjugation.
Embo J., 39, 2020
|
|
7X45
 
 | | Grass carp interferon gamma related | | Descriptor: | Interferon gamma | | Authors: | Wang, J, Zou, J, Zhu, X. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel Dimeric Architecture of an IFN-gamma-Related Cytokine Provides Insights into Subfunctionalization of Type II IFNs in Teleost Fish.
J Immunol., 209, 2022
|
|
4DAD
 
 | |
7DCL
 
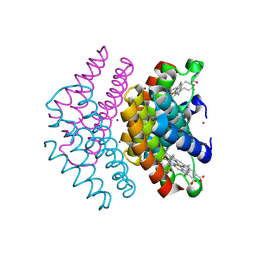 | |
3WAN
 
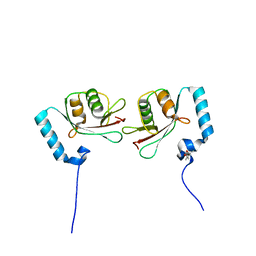 | | Crystal structure of Atg13 LIR-fused human LC3A_2-121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3E4Y
 
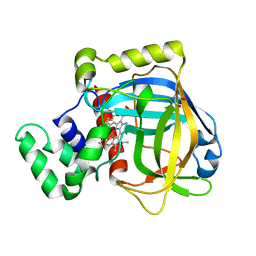 | |
8TLN
 
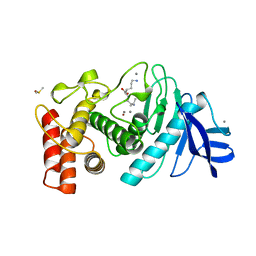 | |
3G3S
 
 | |
4E89
 
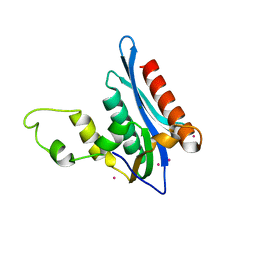 | | Crystal Structure of RnaseH from gammaretrovirus | | Descriptor: | CADMIUM ION, MAGNESIUM ION, RNase H | | Authors: | Kim, J.H, Kim, S.J. | | Deposit date: | 2012-03-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of xenotropic murine leukaemia virus-related virus (XMRV) ribonuclease H
Biosci.Rep., 32, 2012
|
|
8UYL
 
 | | MERS 5' proximal stem-loop 5, conformation 2 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYM
 
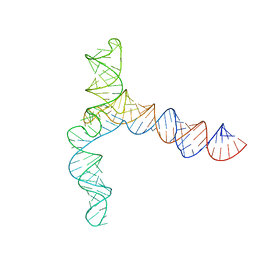 | | MERS 5' proximal stem-loop 5, conformation 3 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4EYZ
 
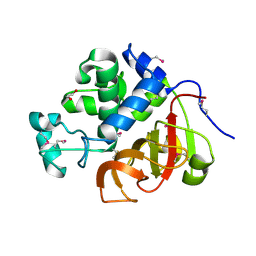 | | Crystal structure of an uncommon cellulosome-related protein module from Ruminococcus flavefaciens that resembles papain-like cysteine peptidases | | Descriptor: | 1,2-ETHANEDIOL, Cellulosome-related protein module from Ruminococcus flavefaciens that resembles papain-like cysteine peptidases | | Authors: | Frolow, F, Voronov-Goldman, M, Bayer, E, Lamed, R. | | Deposit date: | 2012-05-02 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Crystal Structure of an Uncommon Cellulosome-Related Protein Module from Ruminococcus flavefaciens That Resembles Papain-Like Cysteine Peptidases.
Plos One, 8, 2013
|
|
4DXA
 
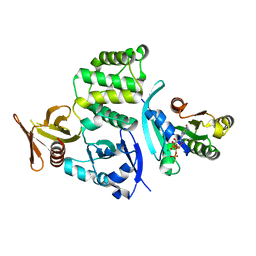 | | Co-crystal structure of Rap1 in complex with KRIT1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Li, X, Zhang, R, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Small G Protein Effector Interaction of Ras-related Protein 1 (Rap1) and Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 287, 2012
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-01-18 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
5Z5O
 
 | | Structure of Pycnonodysostosis disease related I249T mutant of human cathepsin K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Biswas, S, Roy, S. | | Deposit date: | 2018-01-19 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Not all pycnodysostosis-related mutants of human cathepsin K are inactive - crystal structure and biochemical studies of an active mutant I249T.
FEBS J., 285, 2018
|
|
3LLU
 
 | | Crystal structure of the nucleotide-binding domain of Ras-related GTP-binding protein C | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related GTP-binding protein C, ... | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, Crombet, L, Zhong, N, Guan, X, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the nucleotide-binding domain of Ras-related GTP-binding protein C
to be published
|
|
3E4W
 
 | |
3RJ3
 
 | |
