4E7D
 
 | |
6RV8
 
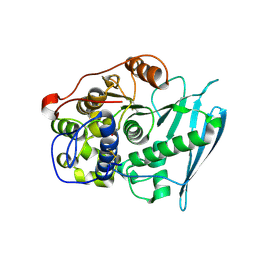 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-Xylitol, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
1JWV
 
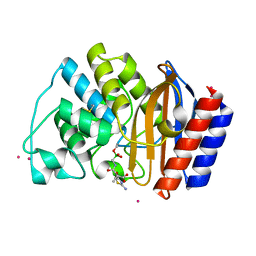 | |
6JKB
 
 | | Crystal structure of metallo-beta-lactamse, NDM-1, in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
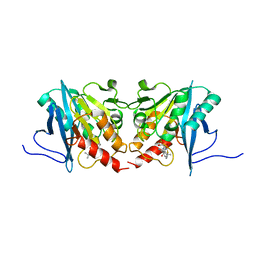
3O5V
 
 | | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A | | Descriptor: | CHLORIDE ION, GLYCEROL, X-PRO dipeptidase | | Authors: | Stein, A.J, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A
To be Published
|
|
7WFF
 
 | | Subcomplexes B,M and L in the Cylic electron transfer supercomplex NDH-PSI from Arabidopsis | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
4LTC
 
 | | Crystal structure of yeast 20S proteasome in complex with enone carmaphycin analogue 6 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(4S,5S,6R)-5-hydroxy-2,6-dimethyloctan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Stein, M, Trivella, D.B.B, Groll, M. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
1ONN
 
 | | IspC apo structure | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase | | Authors: | Steinbacher, S, Kaiser, J, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol
4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria
drug development.
J.BIOL.CHEM., 278, 2003
|
|
6ROH
 
 | | Cryo-EM structure of the autoinhibited Drs2p-Cdc50p | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
4ONW
 
 | | Crystal structure of the catalytic domain of DapE protein from V.cholerea | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, ACETATE ION, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Jedrzejczak, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Dimerization Domain in DapE Enzymes Is required for Catalysis.
Plos One, 9, 2014
|
|
7LUA
 
 | | Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848 SOSIP gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
5SJG
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C4CCN(c1cn(c(n1)CCc3nn2c(ncc(c2n3)C)C)C)C4=O, micromolar IC50=0.039238 | | Descriptor: | 1-(2-{2-[(4S)-5,8-dimethyl[1,2,4]triazolo[1,5-c]pyrimidin-2-yl]ethyl}-1-methyl-1H-imidazol-4-yl)pyrrolidin-2-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
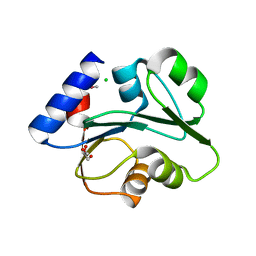
4E7G
 
 | |
1WEG
 
 | | Catalytic Domain Of Muty From Escherichia Coli K142A Mutant | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, IMIDAZOLE, ... | | Authors: | Hitomi, K, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
4OP4
 
 | | Crystal structure of the catalytic domain of DapE protein from V.cholerea in the Zn bound form | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, GLYCEROL, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | The Dimerization Domain in DapE Enzymes Is required for Catalysis.
Plos One, 9, 2014
|
|
4M6L
 
 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and 5,10-dideazatetrahydrofolic acid | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dihydrofolate reductase, N-(4-{2-[(6S)-2-amino-4-oxo-1,4,5,6,7,8-hexahydropyrido[2,3-d]pyrimidin-6-yl]ethyl}benzoyl)-L-glutamic acid, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1P7H
 
 | | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kB element | | Descriptor: | 5'-D(*AP*AP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*A)-3', 5'-D(*TP*TP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | Authors: | Giffin, M.J, Stroud, J.C, Bates, D.L, von Koenig, K.D, Hardin, J, Chen, L. | | Deposit date: | 2003-05-01 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element
Nat.Struct.Biol., 10, 2003
|
|
3WS9
 
 | | Crystal structure of PDE10A in complex with a benzimdazole inhibitor | | Descriptor: | 7-[2-(5-methyl-1-phenyl-1H-benzimidazol-2-yl)ethyl]imidazo[1,5-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Novel benzimidazole derivatives as phosphodiesterase 10A (PDE10A) inhibitors with improved metabolic stability.
Bioorg.Med.Chem., 22, 2014
|
|
1JI2
 
 | | Improved X-ray Structure of Thermoactinomyces vulgaris R-47 alpha-Amylase 2 | | Descriptor: | ALPHA-AMYLASE II, CALCIUM ION | | Authors: | Kamitori, S, Abe, A, Ohtaki, A, Kaji, A, Tonozuka, T, Sakano, Y. | | Deposit date: | 2001-06-28 | | Release date: | 2002-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 alpha-amylase 1 (TVAI) at 1.6 A resolution and alpha-amylase 2 (TVAII) at 2.3 A resolution.
J.Mol.Biol., 318, 2002
|
|
4YFK
 
 | | Escherichia coli RNA polymerase in complex with squaramide compound 8. | | Descriptor: | 3,5-dimethyl-N-{2-[4-(4-methylbenzyl)piperidin-1-yl]-3,4-dioxocyclobut-1-en-1-yl}-1,2-oxazole-4-sulfonamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.571 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4OX9
 
 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmA bound to the 30S ribosomal subunit | | Descriptor: | 16S rRNA, 16S rRNA (adenine(1408)-N(1))-methyltransferase, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Conn, G.L, Dunham, C.M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8035 Å) | | Cite: | Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7LU9
 
 | | Cryo-EM structure of DH851.3 bound to HIV-1 CH505 Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
5SJZ
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH N1(CCOCC1)C(c2c(n(nc2)C)C(Nc4cc3nc(nn3cc4)N5CCCC5)=O)=O, micromolar IC50=0.0008908 | | Descriptor: | 1-methyl-4-(morpholine-4-carbonyl)-N-[(4S)-2-(pyrrolidin-1-yl)[1,2,4]triazolo[1,5-a]pyridin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4MAD
 
 | | Crystal structure of beta-galactosidase C (BgaC) from Bacillus circulans ATCC 31382 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-galactosidase | | Authors: | Kamerke, C, You, D.J, Kanaya, S, Elling, L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of a glycosynthase by the crystal structure of beta-galactosidase from Bacillus circulans (BgaC) and its use for the synthesis of N-acetyllactosamine type 1 glycan structures.
J.Biotechnol., 191, 2014
|
|
3O0T
 
 | | Crystal structure of human phosphoglycerate mutase family member 5 (PGAM5) in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
