6QDV
 
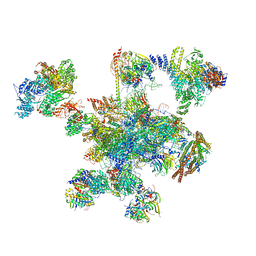 | | Human post-catalytic P complex spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Fica, S.M, Oubridge, C, Wilkinson, M.E, Newman, A.J, Nagai, K. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation.
Science, 363, 2019
|
|
6PUU
 
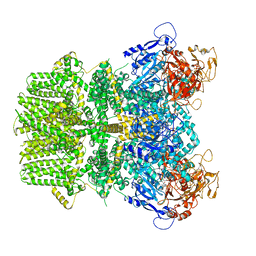 | | Human TRPM2 bound to 8-Br-cADPR and calcium | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
1FRV
 
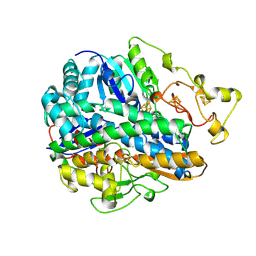 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF NI-FE HYDROGENASE | | Descriptor: | FE3-S4 CLUSTER, HYDRATED FE, HYDROGENASE, ... | | Authors: | Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1996-03-28 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas.
Nature, 373, 1995
|
|
8J0A
 
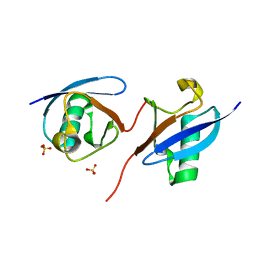 | | Robust design of effective allosteric activator UbV R4 for Rsp5 E3 ligase using the machine-learning tool ProteinMPNN | | Descriptor: | SULFATE ION, Ubiquitin variant R4 | | Authors: | Lin, Y.-F, Hsieh, Y.-J, Kao, H.-W, Ko, T.-P, Wu, K.-P. | | Deposit date: | 2023-04-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Robust Design of Effective Allosteric Activators for Rsp5 E3 Ligase Using the Machine Learning Tool ProteinMPNN.
Acs Synth Biol, 12, 2023
|
|
6Z4R
 
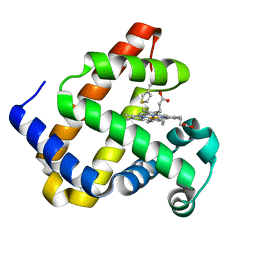 | |
6Z4T
 
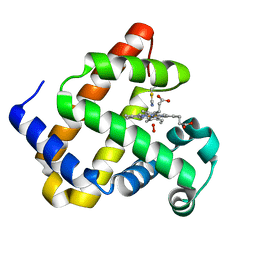 | |
1EV2
 
 | | CRYSTAL STRUCTURE OF FGF2 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 2 (FGFR2) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 2), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 2), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
9DRY
 
 | | CHIP U-box dimer bound to Fab 2F1 | | Descriptor: | 2F1 Fab heavy chain, 2F1 Fab light chain, E3 ubiquitin-protein ligase CHIP | | Authors: | Unnikrishnan, A, Southworth, D. | | Deposit date: | 2024-09-26 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Recombinant antibodies inhibit enzymatic activity of the E3 ubiquitin ligase CHIP via multiple mechanisms.
J.Biol.Chem., 301, 2025
|
|
3NZ7
 
 | |
1XU1
 
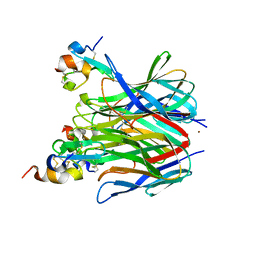 | | The crystal structure of APRIL bound to TACI | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
7QPP
 
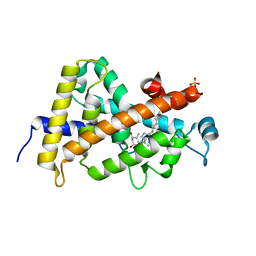 | | High resolution structure of human VDR ligand binding domain in complex with calcitriol | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, SULFATE ION, Vitamin D3 receptor | | Authors: | Rochel, N. | | Deposit date: | 2022-01-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Advances in Vitamin D Receptor Function and Evolution Based on the 3D Structure of the Lamprey Ligand-Binding Domain.
J.Med.Chem., 65, 2022
|
|
6PUO
 
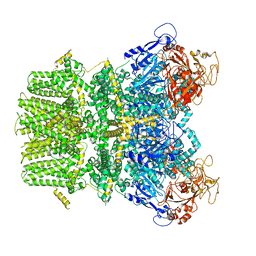 | | Human TRPM2 in the apo state | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
2HUW
 
 | |
9DNT
 
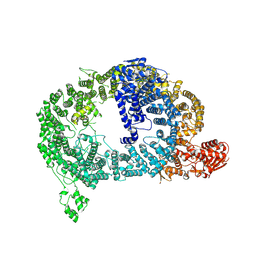 | | Cryo-EM structure of Tom1 (S. cerevisiae) | | Descriptor: | E3 ubiquitin-protein ligase TOM1 | | Authors: | Warner, K.M, Hunkeler, M, Baek, K, Roy Burman, S.S, Fischer, E.S. | | Deposit date: | 2024-09-18 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural ubiquitin contributes to K48 linkage specificity of the HECT ligase Tom1.
Cell Rep, 44, 2025
|
|
1JZL
 
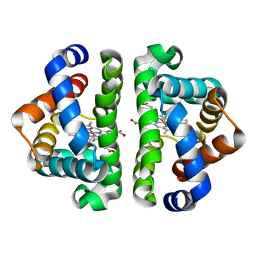 | | Crystal structure of Sapharca inaequivalvis HbI, I114M mutant ligated to carbon monoxide. | | Descriptor: | CARBON MONOXIDE, GLOBIN I - ARK SHELL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-16 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restricting the Ligand-Linked Heme Movement in Scapharca Dimeric Hemoglobin Reveals Tight Coupling between Distal and Proximal
Contributions to Cooperativity.
Biochemistry, 40, 2001
|
|
5MVB
 
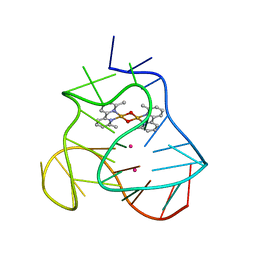 | | Solution structure of a human G-Quadruplex hybrid-2 form in complex with a Gold-ligand. | | Descriptor: | DNA (26-MER), POTASSIUM ION, bis(m2-Oxo)-bis(2-methyl-2,2'-bipyridine)-di-gold(iii) | | Authors: | Wirmer-Bartoschek, J, Jonker, H.R.A, Bendel, L.E, Gruen, T, Bazzicalupi, C, Messori, L, Gratteri, P, Schwalbe, H. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Ligand/Hybrid-2-G-Quadruplex Complex Reveals Rearrangements that Affect Ligand Binding.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5VK4
 
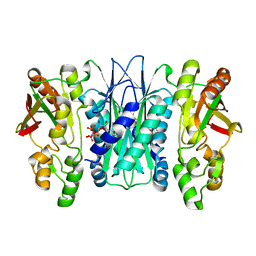 | |
8AT1
 
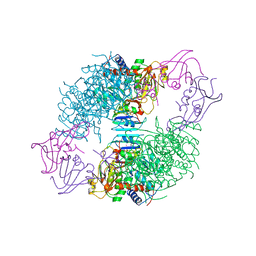 | | CRYSTAL STRUCTURES OF ASPARTATE CARBAMOYLTRANSFERASE LIGATED WITH PHOSPHONOACETAMIDE, MALONATE, AND CTP OR ATP AT 2.8-ANGSTROMS RESOLUTION AND NEUTRAL P*H | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Gouaux, J.E, Stevens, R.C, Lipscomb, W.N. | | Deposit date: | 1989-09-22 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate, and CTP or ATP at 2.8-A resolution and neutral pH.
Biochemistry, 29, 1990
|
|
1XU2
 
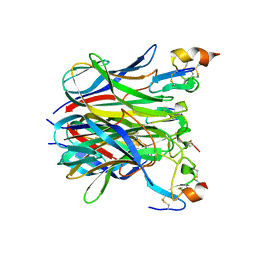 | | The crystal structure of APRIL bound to BCMA | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
3OMO
 
 | | Fragment-Based Design of novel Estrogen Receptor Ligands | | Descriptor: | 2-(trifluoroacetyl)-1,2,3,4-tetrahydroisoquinolin-6-ol, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, van Otterlo, W.A, Rose, R, Fuchs, S, Dominguez Seoane, M, Waldmann, H, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Design and Evaluation of Fragment-Like Estrogen Receptor Tetrahydroisoquinoline Ligands from a Scaffold-Detection Approach.
J.Med.Chem., 54, 2011
|
|
1VWN
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 4.8 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
3OMP
 
 | | Fragment-Based Design of novel Estrogen Receptor Ligands | | Descriptor: | 2-(trifluoroacetyl)-1,2,3,4-tetrahydroisoquinolin-7-ol, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, van Otterlo, W.A, Rose, R, Fuchs, S, Dominguez Seoane, M, Waldmann, H, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design and Evaluation of Fragment-Like Estrogen Receptor Tetrahydroisoquinoline Ligands from a Scaffold-Detection Approach.
J.Med.Chem., 54, 2011
|
|
1VWP
 
 | |
1VWJ
 
 | |
1VWG
 
 | |
