1E8E
 
 | | Solution Structure of Methylophilus methylotrophus Cytochrome c''. Insights into the Structural Basis of Haem-Ligand Detachment | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Brennan, L, Turner, D.L, Fareleira, P, Santos, H. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methylophilus Methylotrophus Cytochrome C": Insights Into the Structural Basis of Haem-Ligand Detachment
J.Mol.Biol., 308, 2001
|
|
2AM9
 
 | | Crystal structure of human androgen receptor ligand binding domain in complex with testosterone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Androgen receptor, GLYCEROL, ... | | Authors: | Pereira de Jesus-Tran, K, Cote, P.-L, Cantin, L, Blanchet, J, Labrie, F, Breton, R. | | Deposit date: | 2005-08-09 | | Release date: | 2006-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity.
Protein Sci., 15, 2006
|
|
3CS4
 
 | | Structure-based design of a superagonist ligand for the vitamin D nuclear receptor | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha)-17-[(2S,4S)-4-(2-hydroxy-2-methylpropyl)-2-methyltetrahydrofuran-2-yl]-9,10-secoandrosta-5,7,10-triene-1,3-diol, Vitamin D3 receptor | | Authors: | Hourai, S, Rodriguez, L.C, Antony, P, Reina-San-Martin, B, Ciesielski, F, Magnier, B.C, Schoonjans, K, Mourino, A, Rochel, N, Moras, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a superagonist ligand for the vitamin d nuclear receptor.
Chem.Biol., 15, 2008
|
|
6TGK
 
 | | Domain swapped E6AP C-lobe dimer | | Descriptor: | ACETATE ION, CALCIUM ION, Ubiquitin-protein ligase E3A | | Authors: | Ries, L.K, Feiler, C, Lowe, L.D, Liess, A.K.L, Lorenz, S. | | Deposit date: | 2019-11-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the catalytic C-lobe of the HECT-type ubiquitin ligase E6AP.
Protein Sci., 29, 2020
|
|
3KIZ
 
 | |
3NYP
 
 | |
6IDE
 
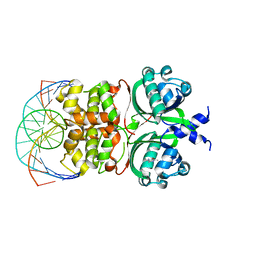 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
3DYB
 
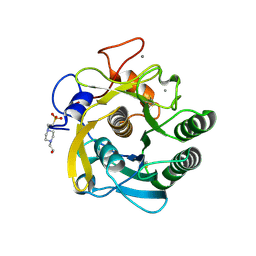 | | proteinase K- digalacturonic acid complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Proteinase K, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A, Cudney, R, Nguyen, C, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | High-resolution structure of proteinase K cocrystallized with digalacturonic acid.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3OMO
 
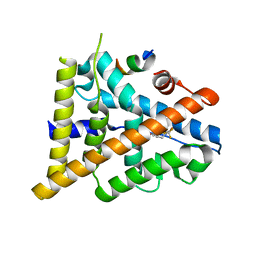 | | Fragment-Based Design of novel Estrogen Receptor Ligands | | Descriptor: | 2-(trifluoroacetyl)-1,2,3,4-tetrahydroisoquinolin-6-ol, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, van Otterlo, W.A, Rose, R, Fuchs, S, Dominguez Seoane, M, Waldmann, H, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Design and Evaluation of Fragment-Like Estrogen Receptor Tetrahydroisoquinoline Ligands from a Scaffold-Detection Approach.
J.Med.Chem., 54, 2011
|
|
6JHU
 
 | | Crystal Structure Of Biotin Protein Ligase From Leishmania Major in complex with Biotinyl-5-AMP | | Descriptor: | BIOTINYL-5-AMP, Biotin/lipoate protein ligase-like protein, SULFATE ION | | Authors: | Rajak, M, Patel, A, Sundd, M. | | Deposit date: | 2019-02-19 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Leishmania major biotin protein ligase forms a unique cross-handshake dimer
Acta Crystallogr.,Sect.D, 77, 2021
|
|
6MKZ
 
 | | Crystal structure of murine 4-1BB/4-1BBL complex | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bitra, A, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the m4-1BB/4-1BBL complex reveals an unusual dimeric ligand that undergoes structural changes upon 4-1BB receptor binding.
J. Biol. Chem., 294, 2019
|
|
7ODU
 
 | | Natural killer cell receptor NKR-P1B from Rattus norvegicus in complex with its cognate ligand Clr-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D11, ... | | Authors: | Skalova, T, Blaha, J, Kalouskova, B, Skorepa, O, Vanek, O, Dohnalek, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Natural killer cell receptor NKR-P1B from Rattus norvegicus in complex with its cognate ligand Clr-11
To Be Published
|
|
3NZ7
 
 | |
3OMQ
 
 | | Fragment-Based Design of novel Estrogen Receptor Ligands | | Descriptor: | 2-[(trifluoromethyl)sulfonyl]-1,2,3,4-tetrahydroisoquinolin-6-ol, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, van Otterlo, W.A, Rose, R, Fuchs, S, Dominguez Seoane, M, Waldmann, H, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design and Evaluation of Fragment-Like Estrogen Receptor Tetrahydroisoquinoline Ligands from a Scaffold-Detection Approach.
J.Med.Chem., 54, 2011
|
|
2ZFX
 
 | | Crystal structure of the rat vitamin D receptor ligand binding domain complexed with YR301 and a synthetic peptide containing the NR2 box of DRIP 205 | | Descriptor: | (2S)-3-{4-[1-ethyl-1-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)propyl]-2-methylphenoxy}propane-1,2-diol, DRIP 205 NR2 box peptide, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2008-01-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of the ligand-binding domain of rat VDR in complex with the nonsecosteroidal vitamin D3 analogue YR301
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3MDO
 
 | |
1UOM
 
 | | The Structure of Estrogen Receptor in Complex with a Selective and Potent Tetrahydroisochiolin Ligand. | | Descriptor: | 2-PHENYL-1-[4-(2-PIPERIDIN-1-YL-ETHOXY)-PHENYL]-1,2,3,4-TETRAHYDRO-ISOQUINOLIN-6-OL, ESTROGEN RECEPTOR | | Authors: | Stark, W, Bischoff, S.F, Buhl, T, Fournier, B, Halleux, C, Kallen, J, Keller, H, Renaud, J. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Estrogen Receptor Modulators: Identification and Structure-Activity Relationships of Potent Eralpha-Selective Tetrahydroisoquinoline Ligands
J.Med.Chem., 46, 2003
|
|
7QPP
 
 | | High resolution structure of human VDR ligand binding domain in complex with calcitriol | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, SULFATE ION, Vitamin D3 receptor | | Authors: | Rochel, N. | | Deposit date: | 2022-01-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Advances in Vitamin D Receptor Function and Evolution Based on the 3D Structure of the Lamprey Ligand-Binding Domain.
J.Med.Chem., 65, 2022
|
|
1OCA
 
 | | HUMAN CYCLOPHILIN A, UNLIGATED, NMR, 20 STRUCTURES | | Descriptor: | CYCLOPHILIN A | | Authors: | Ottiger, M, Zerbe, O, Guntert, P, Wuthrich, K. | | Deposit date: | 1997-07-07 | | Release date: | 1997-11-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution conformation of unligated human cyclophilin A.
J.Mol.Biol., 272, 1997
|
|
6O3F
 
 | |
1FTO
 
 | |
3OMP
 
 | | Fragment-Based Design of novel Estrogen Receptor Ligands | | Descriptor: | 2-(trifluoroacetyl)-1,2,3,4-tetrahydroisoquinolin-7-ol, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, van Otterlo, W.A, Rose, R, Fuchs, S, Dominguez Seoane, M, Waldmann, H, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design and Evaluation of Fragment-Like Estrogen Receptor Tetrahydroisoquinoline Ligands from a Scaffold-Detection Approach.
J.Med.Chem., 54, 2011
|
|
1TP5
 
 | | Crystal structure of PDZ3 domain of PSD-95 protein complexed with a peptide ligand KKETWV | | Descriptor: | LYS-LYS-GLU-THR-TRP-VAL peptide ligand, Presynaptic density protein 95 | | Authors: | Saro, D, Wawrzak, Z, Martin, P, Vickrey, J, Paredes, A, Kovari, L, Spaller, M. | | Deposit date: | 2004-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of the third PDZ domain of PSD-95 protein complexed with KKETWV peptide ligand
To be Published
|
|
1TP3
 
 | | PDZ3 domain of PSD-95 protein complexed with KKETPV peptide ligand | | Descriptor: | KKETPV peptide ligand, Presynaptic density protein 95 | | Authors: | Saro, D, Martin, P, Vickrey, J.F, Griffin, A, Kovari, L.C, Spaller, M.R. | | Deposit date: | 2004-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of the third PDZ domain of PSD-95 protein complexed with KKETPV peptide ligand
To be Published
|
|
1D2U
 
 | | 1.15 A CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS | | Descriptor: | AMMONIA, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Andersen, J.F, Roberts, S.A, Montfort, W.R. | | Deposit date: | 1999-09-28 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
