4I1Z
 
 | | Crystal structure of the monomeric (V948R) form of the gefitinib/erlotinib resistant EGFR kinase domain L858R+T790M | | Descriptor: | Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
2H14
 
 | |
3VBQ
 
 | |
6GC7
 
 | | 50S ribosomal subunit assembly intermediate state 1 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
7W1R
 
 | | Crystal structure of human Suv3 monomer | | Descriptor: | ATP-dependent RNA helicase SUPV3L1, mitochondrial | | Authors: | Jain, M, Yuan, H.S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dimeric assembly of human Suv3 helicase promotes its RNA unwinding function in mitochondrial RNA degradosome for RNA decay.
Protein Sci., 31, 2022
|
|
8D8I
 
 | | Crystal structure of Reverb alpha in complex with synthetic agonist | | Descriptor: | (4S)-6-[([1,1'-biphenyl]-2-yl)oxy]-3-chloro[1,2,4]triazolo[4,3-b]pyridazine, Nuclear receptor corepressor 1, Nuclear receptor subfamily 1 group D member 1 | | Authors: | Ronin, C, Ciesielski, F, Hegazy, L, Burris, P.T. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis of synthetic agonist activation of the nuclear receptor REV-ERB.
Nat Commun, 13, 2022
|
|
5J8Y
 
 | |
5Q04
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(4-bromo-5-chlorothiophen-2-yl)sulfonyl-3-(5-bromo-1,3-thiazol-2-yl)urea | | Descriptor: | 4-bromo-N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-chlorothiophene-2-sulfonamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(4-bromo-5-chlorothiophen-2-yl)sulfonyl-3-(5-bromo-1,3-thiazol-2-yl)urea
To be published
|
|
5F9B
 
 | |
6VZO
 
 | |
6VZM
 
 | |
6VZN
 
 | |
6VZL
 
 | |
1AII
 
 | | ANNEXIN III | | Descriptor: | ANNEXIN III, CALCIUM ION, ETHANOLAMINE, ... | | Authors: | Lewit-Bentley, A, Perron, B. | | Deposit date: | 1996-11-28 | | Release date: | 1997-03-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Can enzymatic activity, or otherwise, be inferred from structural studies of annexin III?
J.Biol.Chem., 272, 1997
|
|
3IUQ
 
 | | apPEP_D622N+PP closed state | | Descriptor: | GLYCEROL, N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
5PZV
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(4-bromopyridin-2-yl)-3-(4-chlorophenyl)sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(4-bromopyridin-2-yl)carbamoyl]-4-chlorobenzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(4-bromopyridin-2-yl)-3-(4-chlorophenyl)sulfonylurea
To be published
|
|
5Q0A
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-cyanopyrazin-2-yl)-3-[3-(difluoromethoxy)phenyl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-cyanopyrazin-2-yl)carbamoyl]-3-(difluoromethoxy)benzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-cyanopyrazin-2-yl)-3-[3-(difluoromethoxy)phenyl]sulfonylurea
To be published
|
|
2AHP
 
 | |
3TQN
 
 | | Structure of the transcriptional regulator of the GntR family, from Coxiella burnetii. | | Descriptor: | Transcriptional regulator, GntR family | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2BZJ
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PIM1 IN COMPLEX WITH A RUTHENIUM ORGANOMETALLIC LIGAND RU3 | | Descriptor: | PROTO-ONCOGENE SERINE THREONINE PROTEIN KINASE PIM1, RUTHENIUM-PYRIDOCARBAZOLE-3 | | Authors: | Debreczeni, J.E, Bullock, A, Knapp, S, von Delft, F, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-08-18 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Human Pim1 in Complex with Ruthenium Organometallic Ligands
To be Published
|
|
2BZK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PIM1 IN COMPLEX WITH AMPPNP AND PIMTIDE | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PIMTIDE, PROTO-ONCOGENE SERINE THREONINE PROTEIN KINASE PIM1 | | Authors: | Debreczeni, J.E, Bullock, A, Knapp, S, von Delft, F, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-08-18 | | Release date: | 2005-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Human Pim1 in Complex with Amppnp and Pimtide
To be Published
|
|
2C3I
 
 | | CRYSTAL STRUCTURE OF HUMAN PIM1 IN COMPLEX WITH IMIDAZOPYRIDAZIN I | | Descriptor: | 1-(3-{6-[(CYCLOPROPYLMETHYL)AMINO]IMIDAZO[1,2-B]PYRIDAZIN-3-YL}PHENYL)ETHANONE, PIMTIDE, PROTO-ONCOGENE SERINE THREONINE PROTEIN KINASE PIM1 | | Authors: | Philippakopoulos, P, Knapp, S, Debreczeni, J, Bullock, A, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Guo, K, Weigelt, J. | | Deposit date: | 2005-10-07 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis Identifies Imidazo[1,2- B]Pyridazines as Pim Kinase Inhibitors with in Vitro Antileukemic Activity.
Cancer Res., 67, 2007
|
|
2BZI
 
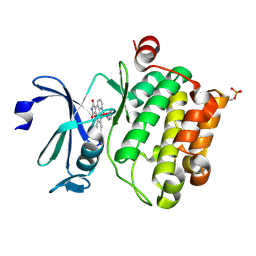 | | CRYSTAL STRUCTURE OF THE HUMAN PIM1 IN COMPLEX WITH A RUTHENIUM ORGANOMETALLIC LIGAND RU2 | | Descriptor: | PROTO-ONCOGENE SERINE THREONINE PROTEIN KINASE PIM1, RU-PYRIDOCARBAZOLE-2 | | Authors: | Debreczeni, J.E, Bullock, A, Knapp, S, von Delft, F, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Pim1 in Complex with Ruthenium Organometallic Ligands
To be Published
|
|
4KRT
 
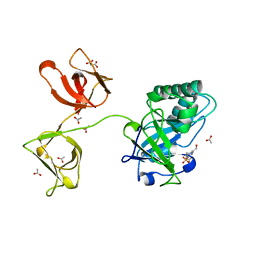 | |
6S8F
 
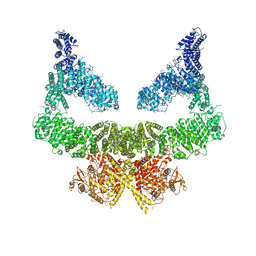 | | Structure of nucleotide-bound Tel1/ATM | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1 | | Authors: | Yates, L.A, Williams, R.M, Ayala, R, Zhang, X. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM Structure of Nucleotide-Bound Tel1ATMUnravels the Molecular Basis of Inhibition and Structural Rationale for Disease-Associated Mutations.
Structure, 28, 2020
|
|
