6SJC
 
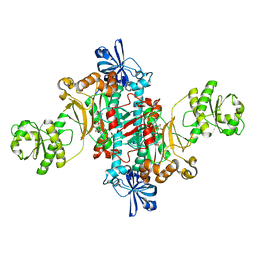 | | Structure of T. thermophilus AspRS in Complex with 5'-O-(N-(L-aspartyl)-sulfamoyl)adenosine | | Descriptor: | 5'-O-(L-alpha-aspartylsulfamoyl)adenosine, Aspartate--tRNA(Asp) ligase | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
1PW6
 
 | | Low Micromolar Small Molecule Inhibitor of IL-2 | | Descriptor: | 2-CYCLOHEXYL-N-(2-{4-[5-(2,3-DICHLORO-PHENYL)-2H-PYRAZOL-3-YL]-PIPERIDIN-1-YL}-2-OXO-ETHYL)-2-GUANIDINO-ACETAMIDE, Interleukin-2, SULFATE ION | | Authors: | Thanos, C.D, Randal, M, Wells, J.A. | | Deposit date: | 2003-06-30 | | Release date: | 2004-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potent small-molecule binding to a dynamic hot spot on IL-2.
J.Am.Chem.Soc., 125, 2003
|
|
2X0V
 
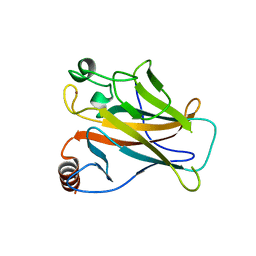 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 4-(trifluoromethyl)benzene-1,2-diamine | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Basse, N, Kaar, J.L, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
3U8L
 
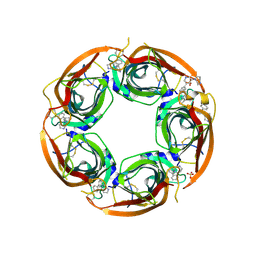 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3570 (1-(5-phenylpyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(5-phenylpyridin-3-yl)-1,4-diazepane, Acetylcholine-binding protein, SULFATE ION | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
3ZSN
 
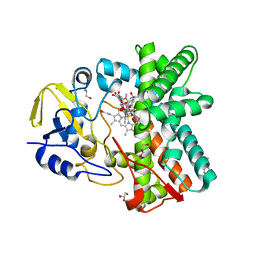 | | Structure of the mixed-function P450 MycG F286A mutant in complex with mycinamicin IV | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Rutaganira, F.U, Anzai, Y, Kato, F, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-06-29 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
4OQP
 
 | |
8Y2R
 
 | | NMR Solution Structure of the 2:1 Coptisine-ATF4-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (5'-D(*TP*AP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*AP*GP*GP*GP*AP*GP*C*(LIG)*(LIG))-3') | | Authors: | Xiao, C.M, Li, Y.P, Liu, Y.S, Dong, R.F, He, X.Y, Lin, Q, Zang, X, Wang, K.B, Xia, Y.Z, Kong, L.Y. | | Deposit date: | 2024-01-27 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | SOLUTION NMR | | Cite: | Overcoming Cancer Persister Cells by Stabilizing the ATF4 Promoter G-quadruplex.
Adv Sci, 11, 2024
|
|
2GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, OXIDIZED GLUTATHIONE COMPLEX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
4F41
 
 | |
1FRQ
 
 | | FERREDOXIN:NADP+ OXIDOREDUCTASE (FERREDOXIN REDUCTASE) MUTANT E312A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, PROTEIN (FERREDOXIN:NADP+ OXIDOREDUCTASE), ... | | Authors: | Aliverti, A, Deng, Z, Ravasi, D, Piubelli, L, Karplus, P.A, Zanetti, G. | | Deposit date: | 1998-10-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the function of the invariant glutamyl residue 312 in spinach ferredoxin-NADP+ reductase.
J.Biol.Chem., 273, 1998
|
|
4DF3
 
 | |
5KZ7
 
 | | Mark2 complex with 7-[(1S)-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(3-pyridylamino)pyrrolo[2,3-d]pyrimidin-6-one | | Descriptor: | 7-[(1~{S})-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(pyridin-3-ylamino)pyrrolo[2,3-d]pyrimidin-6-one, Serine/threonine-protein kinase MARK2 | | Authors: | Su, H.P, Munshi, S.K. | | Deposit date: | 2016-07-23 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure guided design of a series of selective pyrrolopyrimidinone MARK inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6OPV
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPZ
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, I54L, A71V, L76V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
2GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 IN COMPLEX WITH ETHACRYNIC ACID | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A.J, Rossjohn, J, Parker, M.W. | | Deposit date: | 1996-10-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of the human Pi class glutathione transferase P1-1 in complex with the inhibitor ethacrynic acid and its glutathione conjugate.
Biochemistry, 36, 1997
|
|
8AFF
 
 | |
8AFG
 
 | |
3M3E
 
 | |
2QD0
 
 | | Crystal structure of mitoNEET | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH domain-containing protein 1 | | Authors: | Lin, J, Zhou, T, Ye, K, Wang, J. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1URP
 
 | |
4G61
 
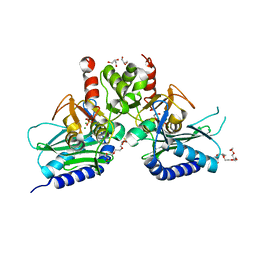 | | Crystal structure of IMPase/NADP phosphatase complexed with Mg2+ and phosphate | | Descriptor: | 1-HYDROXYSULFANYL-4-MERCAPTO-BUTANE-2,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2012-07-18 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
5G4Q
 
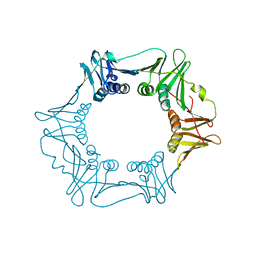 | | H.pylori Beta clamp in complex with 5-chloroisatin | | Descriptor: | 5-chloro-1H-indole-2,3-dione, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening of E. coli beta-clamp Inhibitors Revealed that Few Inhibit Helicobacter pylori More Effectively: Structural and Functional Characterization.
Antibiotics (Basel), 7, 2018
|
|
6CBN
 
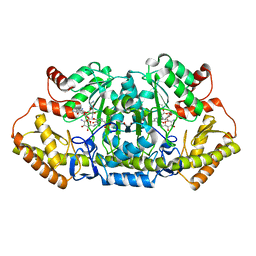 | | x-ray structure of NeoB from streptomyces fradiae in complex with PLP and neomycin (as the external aldimine) at pH 7.5 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-[(3-O-{2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranosyl}-beta-D-ribofuranosyl)oxy]-3-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, Neamine transaminase NeoN | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CEH
 
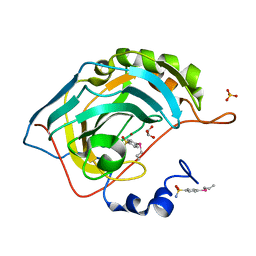 | | Design, Synthesis, X-ray and Biological Activities of Selenides Bearing the Benzenesulfonamide Moiety as New Class of Agents for Prevention of Diabetic Cerebrovascular Pathology | | Descriptor: | 4-[(prop-2-en-1-yl)selanyl]benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Angeli, A, di Cesare Mannelli, L, Trallori, E, Ghelardini, C, Carta, F, Supuran, C.T. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Design, Synthesis, and X-ray of Selenides as New Class of Agents for Prevention of Diabetic Cerebrovascular Pathology.
ACS Med Chem Lett, 9, 2018
|
|
4MMY
 
 | | Integrin AlphaVBeta3 ectodomain bound to the tenth domain of Fibronectin with the IAKGDWND motif | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fibronectin, ... | | Authors: | van Agthoven, J, Xiong, J, Arnaout, M.A. | | Deposit date: | 2013-09-09 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.183 Å) | | Cite: | Structural basis for pure antagonism of integrin alpha V beta 3 by a high-affinity form of fibronectin.
Nat.Struct.Mol.Biol., 21, 2014
|
|
