7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | Authors: | Liu, Z, Liu, S, Yuanzhu, G. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
4AP5
 
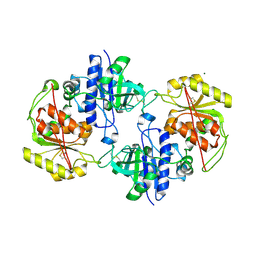 | | Crystal structure of human POFUT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 2, ... | | Authors: | Chen, C, Keusch, J.J, Klein, D, Hess, D, Hofsteenge, J, Gut, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structure of Human Pofut2: Insights Into Thrombospondin Type 1 Repeat Fold and O-Fucosylation.
Embo J., 31, 2012
|
|
5DWI
 
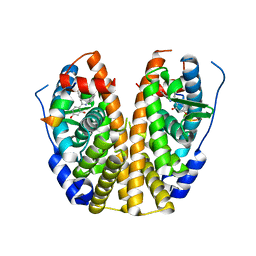 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with a Resorcinyl 2-Chloro-substituted Diaryl-imine analog 4-[(E)-[(2-chlorophenyl)imino](4-hydroxyphenyl)methyl]benzene-1,3-diol | | Descriptor: | 4-[(E)-[(2-chlorophenyl)imino](4-hydroxyphenyl)methyl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6SP8
 
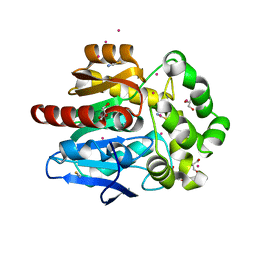 | | Structure of hyperstable haloalkane dehalogenase variant DhaA115 prepared by the 'soak-and-freeze' method under 150 bar of krypton pressure | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Chmelova, K, Markova, K, Damborsky, J, Marek, M. | | Deposit date: | 2019-08-31 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst.
Chem Sci, 11, 2020
|
|
5AGJ
 
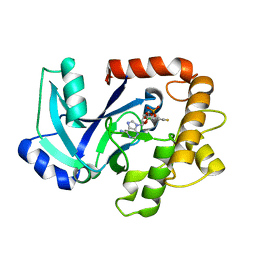 | | Crystal structure of the LeuRS editing domain of Candida albicans in complex with the adduct AN2690-AMP | | Descriptor: | POTENTIAL CYTOSOLIC LEUCYL TRNA SYNTHETASE, [(6-AMINO-9H-PURIN-9-YL)-[5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1-BENZOXABOROLE]-4'YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Zhao, H, Palencia, A, Seiradake, E, Ghaemi, Z, Luthey-Schulten, Z, Cusack, S, Martinis, S.A. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the Resistance Mechanism of a Benzoxaborole Inhibitor Reveals Insight Into the Leucyl-tRNA Synthetase Editing Mechanism.
Acs Chem.Biol., 10, 2015
|
|
5A8C
 
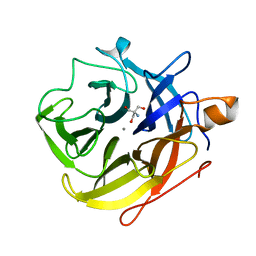 | | The ultra high resolution structure of a novel alpha-L-arabinofuranosidase (CtGH43) from Clostridium thermocellum ATCC 27405 with bound trimethyl N-Oxide (TRS) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6 | | Authors: | Goyal, A, Ahmed, S, Sharma, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-07-14 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Molecular determinants of substrate specificity revealed by the structure of Clostridium thermocellum arabinofuranosidase 43A from glycosyl hydrolase family 43 subfamily 16.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1L5K
 
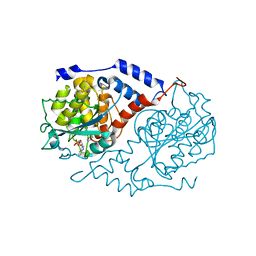 | | Crystal Structure of CobT complexed with N1-(5'-phosphoribosyl)-benzimidazole and nicotinate | | Descriptor: | 1-ALPHA-D-RIBOFURANOSYL-BENZIMIAZOLE-5'-PHOSPHATE, NICOTINIC ACID, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-07 | | Release date: | 2002-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of the L-threonine-O-3-phosphate decarboxylase
(CobD) enzyme from Salmonella enterica: the apo, substrate, and
product-aldimine complexes.
Biochemistry, 41, 2002
|
|
1NMW
 
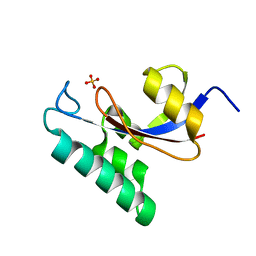 | | Solution structure of the PPIase domain of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
5AB8
 
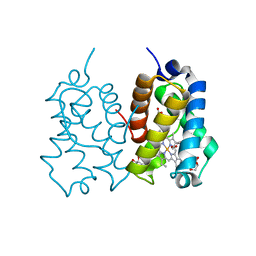 | | High resolution X-ray structure of the N-terminal truncated form (residues 1-11) of Mycobacterium tuberculosis HbN | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Pesce, A, Bustamante, J.P, Bidon-Chanal, A, Boechi, L, Estrin, D.A, Luque, F.J, Sebilo, A, Guertin, M, Bolognesi, M, Ascenzi, P, Nardini, M. | | Deposit date: | 2015-08-04 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The N-Terminal Pre-A Region of Mycobacterium Tuberculosis 2/2Hbn Promotes No-Dioxygenase Activity.
FEBS J., 283, 2016
|
|
7N3T
 
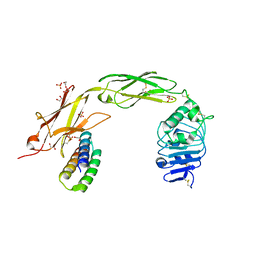 | | TrkA ECD complex with designed miniprotein ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Cao, L, Garcia, K.C. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
3LHN
 
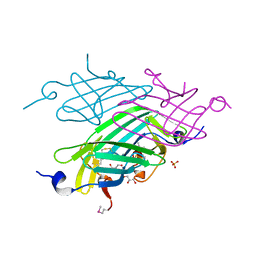 | |
5D25
 
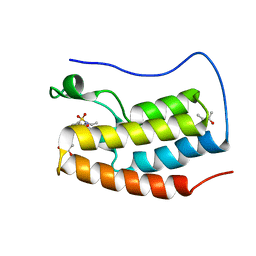 | | First bromodomain of BRD4 bound to inhibitor XD27 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-acetyl-N-[5-(diethylsulfamoyl)-2-hydroxyphenyl]-3,5-dimethyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4, ... | | Authors: | Wohlwend, D, Huegle, M, Gerhardt, S. | | Deposit date: | 2015-08-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D5V
 
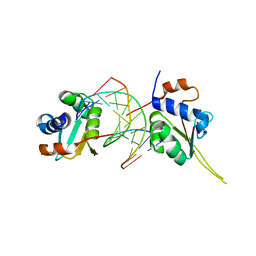 | | Crystal structure of human Hsf1 with Satellite III repeat DNA | | Descriptor: | DNA, Heat shock factor protein 1, MAGNESIUM ION | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5DIT
 
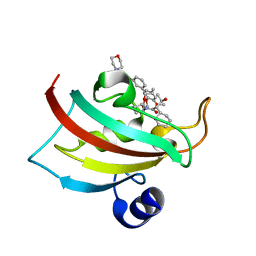 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand (1R)-3-(3,4-dimethoxyphenyl)-1-f3-[2-(morpholin-4-yl)ethoxy]phenylgpropyl(2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Affinity Relationship Analysis of Selective FKBP51 Ligands.
J.Med.Chem., 58, 2015
|
|
9JRP
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound VS-9 | | Descriptor: | 5-[[3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)propanoyl-methyl-amino]methyl]-2-methyl-furan-3-carboxylic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Sun, H, Fang, P. | | Deposit date: | 2024-09-29 | | Release date: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of Aldo-keto reductase 1C3 complexed with compound VS-9
To Be Published
|
|
4RC2
 
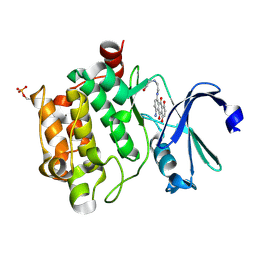 | |
6EUG
 
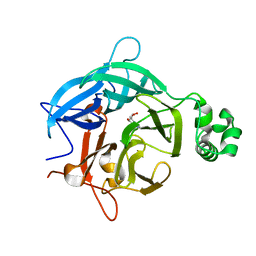 | |
4RCH
 
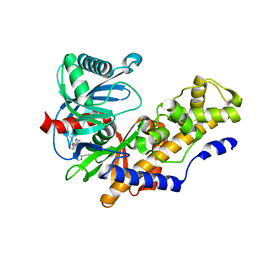 | | Discovery of 2-Pyridyl Ureas as Glucokinase Activators | | Descriptor: | 1-{3-[(2-ethylpyridin-3-yl)oxy]-5-(pyridin-2-ylsulfanyl)pyridin-2-yl}-3-methylurea, Glucokinase, alpha-D-glucopyranose | | Authors: | Voegtli, W, Vigers, G.P.A. | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of 2-pyridylureas as glucokinase activators.
J.Med.Chem., 57, 2014
|
|
1JF7
 
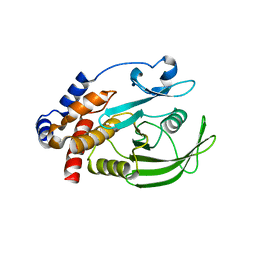 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177836 | | Descriptor: | 5-(2-{2-[(TERT-BUTOXY-HYDROXY-METHYL)-AMINO]-1-HYDROXY-3-PHENYL-PROPYLAMINO}-3-HYDROXY-3-PENTYLAMINO-PROPYL)-2-CARBOXYMETHOXY-BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE 1B | | Authors: | Larsen, S.D, Barf, T, Liljebris, C, May, P.D, Ogg, D, O'Sullivan, T.J, Palazuk, B.J, Schostarez, H.J, Stevens, F.C, Bleasdale, J.E. | | Deposit date: | 2001-06-20 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and biological activity of a novel class of small molecular weight peptidomimetic competitive inhibitors of protein tyrosine phosphatase 1B.
J.Med.Chem., 45, 2002
|
|
6HVA
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with 13 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HVR
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with 16 | | Descriptor: | (2~{S})-~{N}-[(2~{S},3~{R})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-butan-2-yl]-2-[[(2~{R})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
7TN5
 
 | |
7TN6
 
 | |
5TAV
 
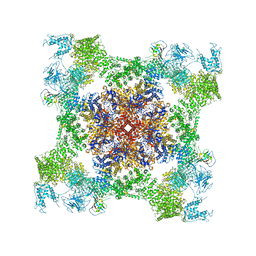 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 4) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
7TN3
 
 | |
