6YVI
 
 | | EED in complex with a cyano-benzofuran | | Descriptor: | 5-fluoranyl-4-[[[8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-yl]amino]methyl]-2,3-dihydro-1-benzofuran-7-carbonitrile, CALCIUM ION, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Read, J.A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Free energy perturbation in the design of EED ligands as inhibitors of polycomb repressive complex 2 (PRC2) methyltransferase.
Bioorg.Med.Chem.Lett., 39, 2021
|
|
7KXT
 
 | | Crystal structure of human EED | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
7MT1
 
 | | Crystal structure of Human Platelet-activating factor acetylhydrolase IB subunit beta (PAFAH1B1) | | Descriptor: | GLYCEROL, Platelet-activating factor acetylhydrolase IB subunit beta, UNKNOWN ATOM OR ION | | Authors: | Hutchinson, A, Seitova, A, Dong, A, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of Human Platelet-activating factor acetylhydrolase IB subunit beta (PAFAH1B1)
To Be Published
|
|
7MSD
 
 | | Structure of EED bound to EEDi-6068 | | Descriptor: | (9aP,12aR)-4-(2,2-difluoropropyl)-12-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,8,11,12a-pentaazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, FORMIC ACID, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
7MSB
 
 | | Structure of EED bound to EEDi-4259 | | Descriptor: | (9aM,12aS)-12-{[(5-fluoro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,11,12a-tetraazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
7KLJ
 
 | | Crystal structure of the WD-repeat domain of human KIF21A | | Descriptor: | 1,2-ETHANEDIOL, Isoform 2 of Kinesin-like protein KIF21A, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Loppnau, P, Hutchinson, A, Seitova, A, Edwards, A.M, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-10-30 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of the WD-repeat domain of human KIF21A
To be Published
|
|
7CFP
 
 | |
7CFQ
 
 | | Crystal structure of WDR5 in complex with H3K4me3Q5ser peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, H3K4me3Q5ser peptide, ... | | Authors: | Zhao, J, Zhang, X, Zang, J. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the recognition of histone H3Q5 serotonylation by WDR5.
Sci Adv, 7, 2021
|
|
7DH6
 
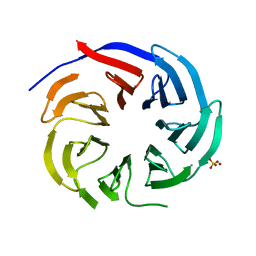 | | Crystal structure of PLRG1 | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Pleiotropic regulator 1, ... | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2020-11-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Crystal structure of the WD40 domain of human PLRG1.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7DNO
 
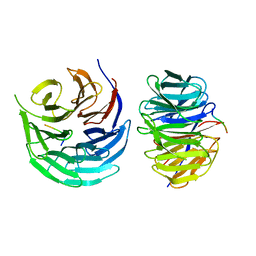 | | Characterization of Peptide Ligands Against WDR5 Isolated Using Phage Display Technique | | Descriptor: | CYS-ARG-THR-LEU-PRO-PHE, WD repeat-containing protein 5 | | Authors: | Cao, J, Cao, D, Xiong, B, Li, Y, Fan, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5.
Molecules, 26, 2021
|
|
7AXU
 
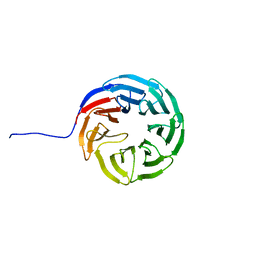 | |
7AXQ
 
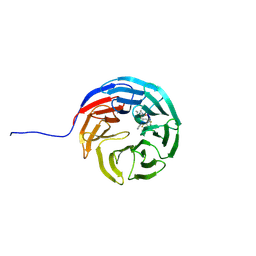 | |
7AXX
 
 | |
7AXP
 
 | |
7AXS
 
 | |
7BID
 
 | | Crystal structure of v31WRAP-T, a 7-bladed designer protein | | Descriptor: | v31WRAP-T | | Authors: | Laier, I, Mylemans, B, Lee, X.Y, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIF
 
 | | Crystal structure of v22WRAP-T, a 7-bladed designer protein | | Descriptor: | v22WRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIG
 
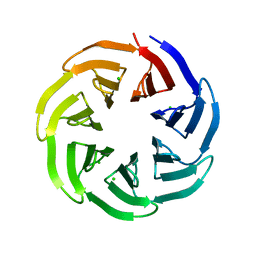 | | Crystal structure of v13WRAP-T, a 7-bladed designer protein | | Descriptor: | CHLORIDE ION, v13WRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIE
 
 | | Crystal structure of nvWrap-T, a 7-bladed symmetric propeller | | Descriptor: | CITRIC ACID, nvWRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BED
 
 | |
7BCY
 
 | |
8Q1N
 
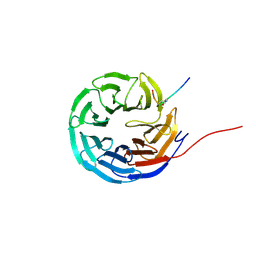 | | Cyclic peptide binder of the WBM-site of WDR5 | | Descriptor: | Cyclic peptide inhibitor, WD repeat-containing protein 5 | | Authors: | Schmeing, S, Chang, J.Y, t Hart, P, Gasper, R. | | Deposit date: | 2023-08-01 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | Macrocyclic peptides as inhibitors of WDR5-lncRNA interactions.
Chem.Commun.(Camb.), 59, 2023
|
|
8KB9
 
 | |
7SI5
 
 | | CRYSTAL STRUCTURE OF EED WITH MRTX-1919 | | Descriptor: | (4R)-8-(1,3-dimethyl-1H-pyrazol-5-yl)-5-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}imidazo[1,2-c]pyrimidine-2-carbonitrile, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Gunn, R.J, Burns, A.C, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | CRYSTAL STRUCTURE OF EED WITH MRTX-1919
TO BE PUBLISHED
|
|
7SI4
 
 | | CRYSTAL STRUCTURE OF EED WITH MRTX-2219 | | Descriptor: | (4S)-8-{4-[(dimethylamino)methyl]-2-methylphenyl}-5-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}imidazo[1,2-c]pyrimidine-2-carbonitrile, FORMIC ACID, Polycomb protein EED | | Authors: | Gunn, R.J, Burns, A.C, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF EED WITH MRTX-2219
TO BE PUBLISHED
|
|
