1PVH
 
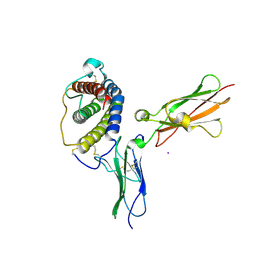 | | Crystal structure of leukemia inhibitory factor in complex with gp130 | | Descriptor: | IODIDE ION, Interleukin-6 receptor beta chain, Leukemia inhibitory factor | | Authors: | Boulanger, M.J, Bankovich, A.J, Kortemme, T, Baker, D, Garcia, K.C. | | Deposit date: | 2003-06-27 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130.
Mol.Cell, 12, 2003
|
|
3FL7
 
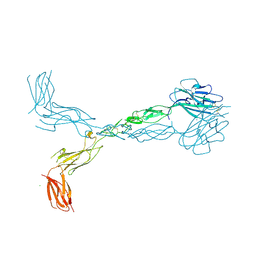 | | Crystal structure of the human ephrin A2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ephrin receptor, ... | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1P9M
 
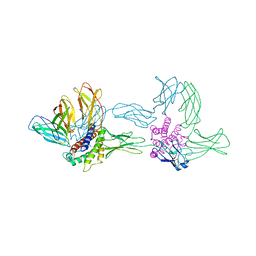 | | Crystal structure of the hexameric human IL-6/IL-6 alpha receptor/gp130 complex | | Descriptor: | Interleukin-6, Interleukin-6 receptor alpha chain, Interleukin-6 receptor beta chain | | Authors: | Boulanger, M.J, Chow, D.C, Brevnova, E.E, Garcia, K.C. | | Deposit date: | 2003-05-12 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Hexameric Structure and Assembly of the Interleukin-6/IL-6 alpha-Receptor/gp130 Complex
Science, 300, 2003
|
|
2Q7N
 
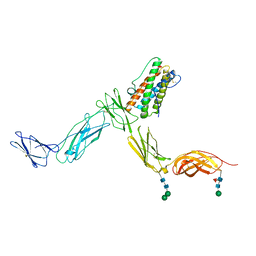 | | Crystal structure of Leukemia inhibitory factor in complex with LIF receptor (domains 1-5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huyton, T, Zhang, J.G, Nicola, N.A, Garrett, T.P.J. | | Deposit date: | 2007-06-07 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
8Q6T
 
 | |
8PMU
 
 | |
5FMV
 
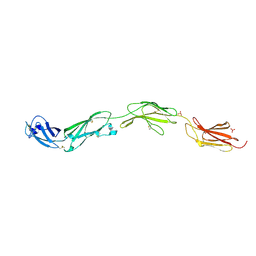 | | Crystal structure of human CD45 extracellular region, domains d1-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C, SULFATE ION | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FM5
 
 | |
6IAA
 
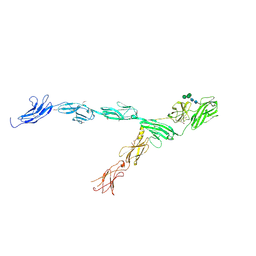 | | hRobo2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Roundabout homolog 2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Barak, R, Isupov, N.M, Opatowsky, Y. | | Deposit date: | 2018-11-26 | | Release date: | 2019-03-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Principles in Robo Activation and Auto-inhibition.
Cell, 177, 2019
|
|
8QY4
 
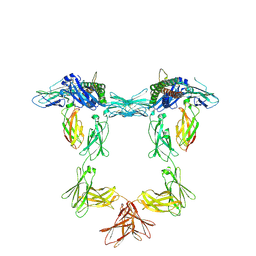 | | Structure of interleukin 11 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure of interleukin 11 (gp130 P496L mutant).
To Be Published
|
|
8QY6
 
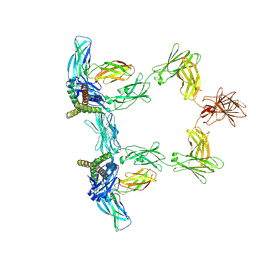 | | Structure of interleukin 6 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of interleukin 6 (gp130 P496L mutant).
To Be Published
|
|
8QY5
 
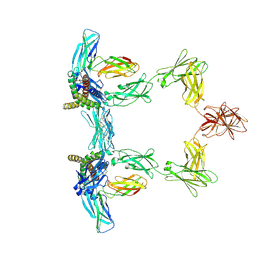 | | Structure of interleukin 6. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of interleukin 6.
To Be Published
|
|
8PN5
 
 | |
8PN3
 
 | |
6M76
 
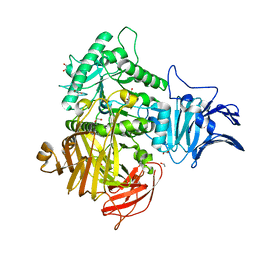 | | GH31 alpha-N-acetylgalactosaminidase from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, LPXTG-motif cell wall anchor domain protein | | Authors: | Miyazaki, T. | | Deposit date: | 2020-03-17 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the Enterococcus faecalis alpha-N-acetylgalactosaminidase, a member of the glycoside hydrolase family 31.
Febs Lett., 594, 2020
|
|
6M77
 
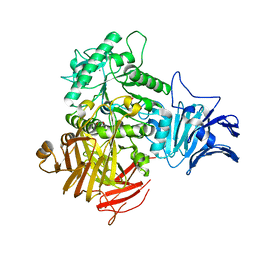 | |
5I99
 
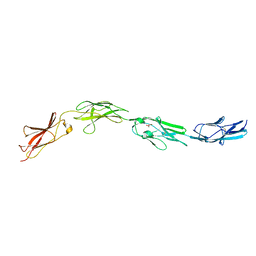 | | Crystal structure of mouse CNTN3 Ig5-Fn2 domains | | Descriptor: | Contactin-3, GLYCEROL | | Authors: | Nikolaienko, R.M, Bouyain, S. | | Deposit date: | 2016-02-19 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Interactions Between Contactin Family Members and Protein-tyrosine Phosphatase Receptor Type G in Neural Tissues.
J.Biol.Chem., 291, 2016
|
|
5J7C
 
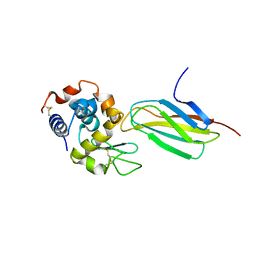 | |
6MOH
 
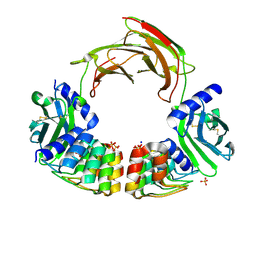 | | Dimeric DARPin C_R3 complex with EpoR | | Descriptor: | Dimeric DARPin E2C (C_R3), Erythropoietin receptor, PHOSPHATE ION, ... | | Authors: | Jude, K.M, Mohan, K, Garcia, K.C. | | Deposit date: | 2018-10-04 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
6MOI
 
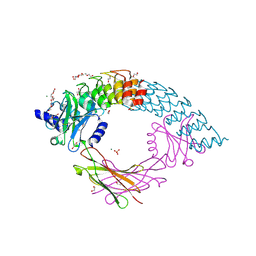 | | Dimeric DARPin C_angle_R5 complex with EpoR | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimeric DARPing CCR5 (C_angle_R5), ... | | Authors: | Jude, K.M, Mohan, K, Garcia, K.C, Guo, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
7SL2
 
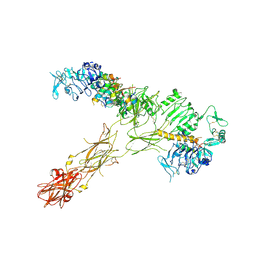 | |
7SL6
 
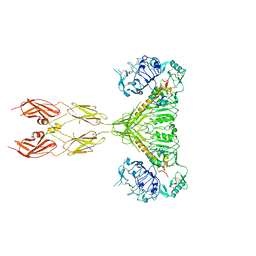 | |
7SL3
 
 | |
7SL4
 
 | |
7SL7
 
 | |
