3SS2
 
 | | Neutron structure of perdeuterated rubredoxin using 48 hours 3rd pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Blakeley, M.P, Weiss, K.L, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-07-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1XJX
 
 | | The crystal structures of the DNA binding sites of the RUNX1 transcription factor | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*GP*GP*TP*C)-3', 5'-D(*TP*GP*AP*CP*CP*GP*CP*AP*G)-3' | | Authors: | Kitayner, M, Rozenberg, H, Rabinovich, D, Shakked, Z. | | Deposit date: | 2004-09-26 | | Release date: | 2005-03-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the DNA-binding site of Runt-domain transcription regulators.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3TAO
 
 | | Structure of Mycobacterium tuberculosis triosephosphate isomerase bound to phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Connor, S.E, Capodagli, G.C, Deaton, M.K, Pegan, S.D. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of Mycobacterium tuberculosis triosephosphate isomerase.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2PS1
 
 | | S. cerevisiae orotate phosphoribosyltransferase complexed with orotic acid and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, OROTIC ACID, ... | | Authors: | Gonzalez-Segura, L, Hurley, T.D, McClard, R.W. | | Deposit date: | 2007-05-04 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ternary complex formation and induced asymmetry in orotate phosphoribosyltransferase.
Biochemistry, 46, 2007
|
|
2Q71
 
 | | Uroporphyrinogen Decarboxylase G168R single mutant enzyme in complex with coproporphyrinogen-III | | Descriptor: | COPROPORPHYRINOGEN III, Uroporphyrinogen decarboxylase | | Authors: | Phillips, J.D, Whitby, F.G, Stadtmueller, B.M, Edwards, C.Q, Hill, C.P, Kushner, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two Novel Uropophyrinogen Decarboxylase (URO-D) Mutations Causing Hepatoerythropoietic Porphyria (HEP)
Transl.Res., 149, 2007
|
|
3FVE
 
 | | Crystal structure of diaminopimelate epimerase Mycobacterium tuberculosis DapF | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Diaminopimelate epimerase, GLYCEROL | | Authors: | Usha, V, Dover, L.G, Roper, D.I, Futterer, K, Besra, G.S. | | Deposit date: | 2009-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the diaminopimelate epimerase DapF from Mycobacterium tuberculosis
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3AUQ
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with Yne-diene type analog of active 14-epi-2alpha-methyl-19-norvitamin D3 | | Descriptor: | (1R,2S,3R)-5-[2-[(1R,3aS,7aR)-1-[(2R)-6-hydroxy-6-methyl-heptan-2-yl]-7a-methyl-1,2,3,3a,6,7-hexahydroinden-4-yl]ethynyl]-2-methyl-cyclohex-4-ene-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Development of 14-epi-19-nortachysterol and its unprecedented binding configuration for the human vitamin D receptor
J.Am.Chem.Soc., 133, 2011
|
|
3AUR
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with Yne-diene type analog of active 14-epi-2beta-methyl-19-norvitamin D3 | | Descriptor: | (1R,2S,3R)-5-[2-[(1R,3aS,7aR)-1-[(2R)-6-hydroxy-6-methyl-heptan-2-yl]-7a-methyl-1,2,3,3a,6,7-hexahydroinden-4-yl]ethynyl]-2-methyl-cyclohex-4-ene-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Development of 14-epi-19-nortachysterol and its unprecedented binding configuration for the human vitamin D receptor
J.Am.Chem.Soc., 133, 2011
|
|
2CNB
 
 | | Trypanosoma brucei UDP-galactose-4-epimerase in ternary complex with NAD and the substrate analogue UDP-4-deoxy-4-fluoro-alpha-D-galactose | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GALACTOSE-4-EPIMERASE, URIDINE-5'-DIPHOSPHATE-4-DEOXY-4-FLUORO-ALPHA-D-GALACTOSE | | Authors: | Alphey, M.S, Ferguson, M.A.J, Hunter, W.N. | | Deposit date: | 2006-05-18 | | Release date: | 2006-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trypanosoma Brucei Udp-Galactose-4-Epimerase in Ternary Complex with Nad+ and the Substrate Analogue Udp-4-Deoxy-4-Fluoro-Alpha-D-Galactose
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3RYG
 
 | | 128 hours neutron structure of perdeuterated rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2CIQ
 
 | | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HEXOSE-6-PHOSPHATE MUTAROTASE, ... | | Authors: | Graille, M, Baltaze, J.-P, Leulliot, N, Liger, D, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2006-03-24 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
J. Biol. Chem., 281, 2006
|
|
2Q5R
 
 | | Structure of apo Staphylococcus aureus D-tagatose-6-phosphate kinase | | Descriptor: | Tagatose-6-phosphate kinase | | Authors: | McGrath, T.E, Soloveychik, M, Romanov, V, Thambipillai, D, Dharamsi, A, Virag, C, Domagala, M, Pai, E.F, Edwards, A.M, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apo Staphylococcus aureus D-tagatose-6-phosphate kinase
TO BE PUBLISHED
|
|
1VS0
 
 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | Authors: | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-27 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
2I5Y
 
 | | Crystal structure of CD4M47, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 GP120 envelope glycoprotein and anti-HIV-1 antibody 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 17B Heavy chain, Antibody 17B Light chain, ... | | Authors: | Huang, C.-C, Kwong, P.D. | | Deposit date: | 2006-08-26 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Combinatorial optimization of a CD4-mimetic miniprotein and cocrystal structures with HIV-1 gp120 envelope glycoprotein.
J.Mol.Biol., 382, 2008
|
|
4F3S
 
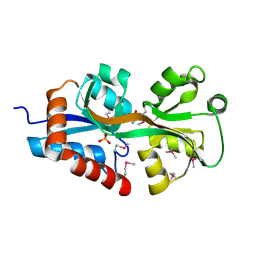 | | Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica | | Descriptor: | D-ALANINE, GLYCINE, PHOSPHATE ION, ... | | Authors: | Agarwal, R, Chamala, S, Evans, B, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Foti, R, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica
To be Published
|
|
2I60
 
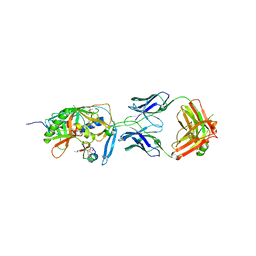 | | Crystal structure of [Phe23]M47, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 GP120 envelope glycoprotein and anti-HIV-1 antibody 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B HEAVY CHAIN, ANTIBODY 17B LIGHT CHAIN, ... | | Authors: | Huang, C.-C, Kwong, P.D. | | Deposit date: | 2006-08-26 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combinatorial optimization of a CD4-mimetic miniprotein and cocrystal structures with HIV-1 gp120 envelope glycoprotein.
J.Mol.Biol., 382, 2008
|
|
2EVS
 
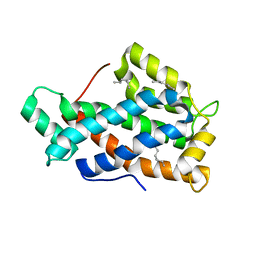 | | Crystal structure of human Glycolipid Transfer Protein complexed with n-hexyl-beta-D-glucoside | | Descriptor: | DECANE, Glycolipid transfer protein, HEXANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2Q6Z
 
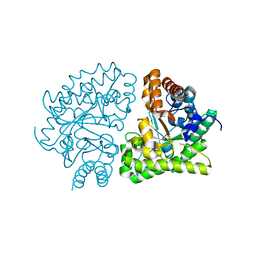 | | Uroporphyrinogen Decarboxylase G168R single mutant apo-enzyme | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Phillips, J.D, Whitby, F.G, Stadtmueller, B.M, Edwards, C.Q, Hill, C.P, Kushner, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two novel uroporphyrinogen decarboxylase (URO-D) mutations causing hepatoerythropoietic porphyria (HEP).
Transl.Res., 149, 2007
|
|
3S2V
 
 | | Crystal Structure of the Ligand Binding Domain of GluK1 in Complex with an Antagonist (S)-1-(2'-Amino-2'-carboxyethyl)-3-[(2-carboxythien-3-yl)methyl]thieno[3,4-d]pyrimidin-2,4-dione at 2.5 A Resolution | | Descriptor: | (S)-1-(2'-AMINO-2'-CARBOXYETHYL)-3-[(2-CARBOXYTHIEN-3-YL)METHYL]THIENO[3,4-D]PYRIMIDIN-2,4-DIONE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective kainate receptor (GluK1) ligands structurally based upon 1H-cyclopentapyrimidin-2,4(1H,3H)-dione: synthesis, molecular modeling, and pharmacological and biostructural characterization.
J.Med.Chem., 54, 2011
|
|
2O4F
 
 | | Structure of a parallel-stranded guanine tetraplex crystallised with monovalent ions | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*T)-3', LITHIUM ION, SODIUM ION | | Authors: | Creze, C, Rinaldi, B, Haser, R, Bouvet, P, Gouet, P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a d(TGGGGT) quadruplex crystallized in the presence of Li+ ions.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2FEQ
 
 | | orally active thrombin inhibitors | | Descriptor: | Decapeptide Hirudin Analogue, N-(CARBOXYMETHYL)-3-CYCLOHEXYL-D-ALANYL-N-({4-[(E)-AMINO(IMINO)METHYL]-1,3-THIAZOL-2-YL}METHYL)-L-PROLINAMIDE, Thrombin heavy chain, ... | | Authors: | Mack, H, Baucke, D, Hornberger, W, Lange, U.E.W, Hoeffken, H.W. | | Deposit date: | 2005-12-16 | | Release date: | 2006-08-08 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Orally active thrombin inhibitors. Part 1: optimization of the P1-moiety
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1DFG
 
 | | X-RAY STRUCTURE OF ESCHERICHIA COLI ENOYL REDUCTASE WITH BOUND NAD AND BENZO-DIAZABORINE | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ENOYL ACYL CARRIER PROTEIN REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baldock, C, Rafferty, J.B, Rice, D.W. | | Deposit date: | 1997-01-16 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mechanism of drug action revealed by structural studies of enoyl reductase.
Science, 274, 1996
|
|
1U64
 
 | | The Solution Structure of d(G3T4G4)2 | | Descriptor: | 5'-D(*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3' | | Authors: | Sket, P, Crnugelj, M, Plavec, J. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | d(G3T4G4) forms unusual dimeric G-quadruplex structure with the same general fold in the presence of K+, Na+ or NH4+ ions.
Bioorg.Med.Chem., 12, 2004
|
|
3RZ6
 
 | | Neutron structure of perdeuterated rubredoxin using 40 hours 1st pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1S23
 
 | | Crystal Structure Analysis of the B-DNA Decamer CGCAATTGCG | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*TP*TP*GP*CP*G)-3', COBALT (II) ION | | Authors: | Valls, N, Wright, G, Steiner, R.A, Murshudov, G.N, Subirana, J.A. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA variability in five crystal structures of d(CGCAATTGCG).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
