3AVP
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with Pantothenol | | Descriptor: | (2S)-2,4-dihydroxy-N-(3-hydroxypropyl)-3,3-dimethylbutanamide, CITRATE ANION, GLYCEROL, ... | | Authors: | Chetnani, B, Kumar, P, Abhinav, K.V, Chhibber, M, Surolia, A, Vijayan, M. | | Deposit date: | 2011-03-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Location and conformation of pantothenate and its derivatives in Mycobacterium tuberculosis pantothenate kinase: insights into enzyme action
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1YYD
 
 | | High Resolution Crystal Structure of Manganese Peroxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Sundaramoorthy, M, Youngs, H.L, Gold, M.H, Poulos, T.L. | | Deposit date: | 2005-02-24 | | Release date: | 2005-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Resolution Crystal Structure of Manganese Peroxidase: Substrate and Inhibitor Complexes.
Biochemistry, 44, 2005
|
|
2E4V
 
 | | Crystal structure of the extracellular region of the group II metabotropic glutamate receptor complexed with DCG-IV | | Descriptor: | (1R,2R)-3-[(S)-amino(carboxy)methyl]cyclopropane-1,2-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Muto, T, Tsuchiya, D, Morikawa, K, Jingami, H. | | Deposit date: | 2006-12-17 | | Release date: | 2007-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the extracellular regions of the group II/III metabotropic glutamate receptors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3JYJ
 
 | | Structure-Based Design of Novel PIN1 Inhibitors (II) | | Descriptor: | (2R,4E)-2-[(naphthalen-2-ylcarbonyl)amino]-5-phenylpent-4-enoic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2009-09-21 | | Release date: | 2010-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (II).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1Z3U
 
 | |
1SVH
 
 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 8 | | Descriptor: | (3R,4S)-N-(4-{TRANS-2-[4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-PHENYL]-VINYL}-AZEPAN-3-YL)-ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
1N87
 
 | |
1MMU
 
 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-glucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, beta-D-glucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
3QVD
 
 | | Exposure of rubrerythrin from Pyrococcus furiosus to peroxide, fifteen second time point. | | Descriptor: | FE (II) ION, FE (III) ION, HYDROGEN PEROXIDE, ... | | Authors: | Dillard, B.D, Demick, J.M, Adams, M.W.W, Lanzilotta, W.N. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A cryo-crystallographic time course for peroxide reduction by rubrerythrin from Pyrococcus furiosus.
J.Biol.Inorg.Chem., 16, 2011
|
|
3EHU
 
 | |
5UJJ
 
 | |
1M6O
 
 | | Crystal Structure of HLA B*4402 in complex with HLA DPA*0201 peptide | | Descriptor: | Beta-2-microglobulin, HLA DPA*0201 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N.A, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-07-17 | | Release date: | 2003-09-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
6FZJ
 
 | | PPAR gamma mutant complex | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CHLORIDE ION, IODIDE ION, ... | | Authors: | Rochel, N. | | Deposit date: | 2018-03-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
1MMZ
 
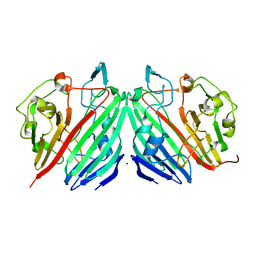 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with L-arabinose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, beta-L-arabinopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, R.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1MMX
 
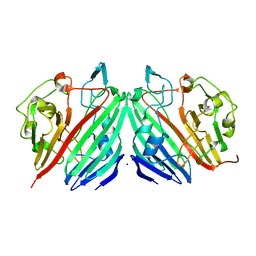 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-fucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, alpha-L-fucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
7KA0
 
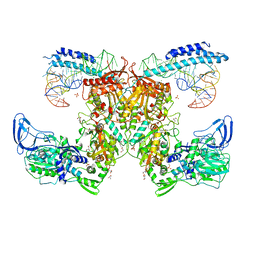 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and phenylalanine | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Wower, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
7KAB
 
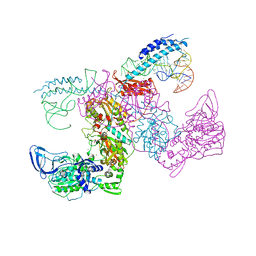 | | M. tuberculosis PheRS complex with cognate precursor tRNA and phenylalanine | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHENYLALANINE, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Wower, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
2A2I
 
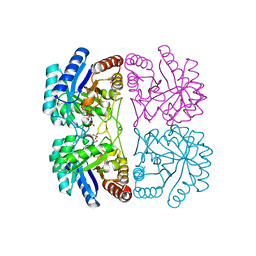 | | Aquifex aeolicus KDO8PS in complex with PEP, A5P, Zn2+ | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, PHOSPHOENOLPYRUVATE, ... | | Authors: | Kona, F, Xu, X, Lu, J, Martin, P, Gatti, D.L. | | Deposit date: | 2005-06-22 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
3FP5
 
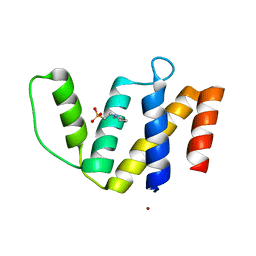 | | Crystal structure of ACBP from Moniliophthora perniciosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acyl-CoA Binding Protein, ZINC ION | | Authors: | Monzani, P.S, Pereira, H.M, Melo, F.A, Meirelles, F.V, Oliva, G, Cascardo, J.C.M. | | Deposit date: | 2009-01-04 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A new topology of ACBP from Moniliophthora perniciosa.
Biochim.Biophys.Acta, 1804, 2010
|
|
6C0Y
 
 | |
2A32
 
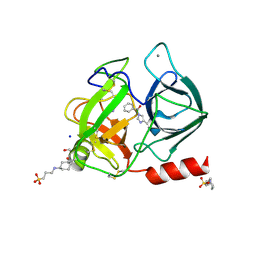 | | Trypsin in complex with benzene boronic acid | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, BORATE ION, CALCIUM ION, ... | | Authors: | Transue, T.R, Gabel, S.A, London, R.E. | | Deposit date: | 2005-06-23 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NMR and crystallographic characterization of adventitious borate binding by trypsin.
Bioconjug.Chem., 17, 2006
|
|
1STN
 
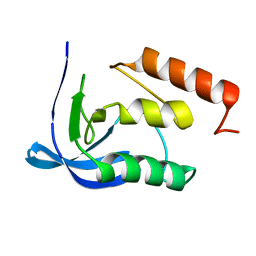 | |
3ERG
 
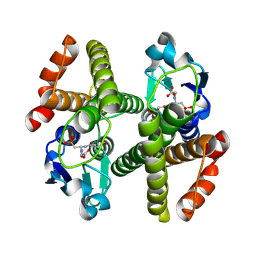 | | Crystal structure of Gtt2 from Saccharomyces cerevisiae in complex with glutathione sulfnate | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-transferase 2 | | Authors: | Ma, X.X, Jiang, Y.L, He, Y.X, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2008-10-02 | | Release date: | 2009-10-13 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family.
Embo Rep., 10, 2009
|
|
4TNW
 
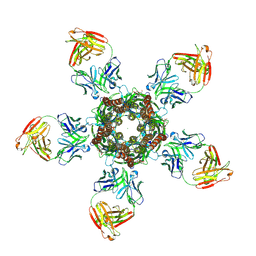 | | C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab and POPC in a lipid-modulated conformation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avermectin-sensitive glutamate-gated chloride channel GluCl alpha, ... | | Authors: | Althoff, T, Hibbs, R.E, Banerjee, S, Gouaux, E. | | Deposit date: | 2014-06-05 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors.
Nature, 512, 2014
|
|
2PR4
 
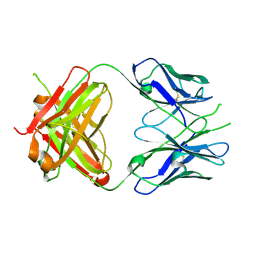 | | Crystal Structure of Fab' from the HIV-1 Neutralizing Antibody 2F5 | | Descriptor: | nmAb 2F5 Fab' Heavy Chain, nmAb 2F5 Fab' light Chain | | Authors: | Bryson, S, Julien, J.-P, Pai, E.F. | | Deposit date: | 2007-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
