6I0I
 
 | |
6I0K
 
 | | Structure of quinolinate synthase in complex with 4-mercaptophthalic acid | | Descriptor: | 4-mercaptoidenecyclohexa-2,5-diene-1,2-dicarboxylic acid, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design of specific inhibitors of quinolinate synthase based on [4Fe-4S] cluster coordination.
Chem.Commun.(Camb.), 55, 2019
|
|
8A0P
 
 | | Crystal structure of poplar glutathione transferase U20 in complex with morin | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, CHLORIDE ION, Glutathione transferase | | Authors: | Didierjean, C, Favier, F. | | Deposit date: | 2022-05-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
7ZS3
 
 | | Crystal structure of poplar glutathione transferase U19 | | Descriptor: | ACETATE ION, Glutathione transferase | | Authors: | Didierjean, C, Favier, F. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
7ZVU
 
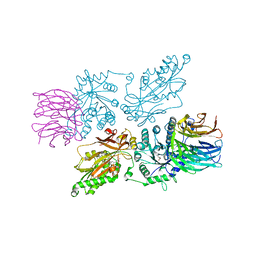 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, Methylosome protein 50, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7ZY8
 
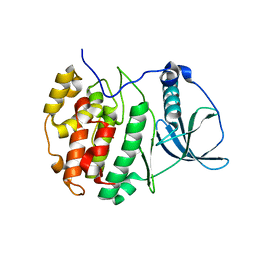 | | Crystal structure of compound 2 bound to CK2alpha | | Descriptor: | 3-[3,5-bis(chloranyl)phenyl]propan-1-amine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Fusco, C, Atkinson, E, Rossmann, M, Francis, N, Iegre, J, Hyvonen, M, Spring, D. | | Deposit date: | 2022-05-24 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
8RZV
 
 | |
7ZY0
 
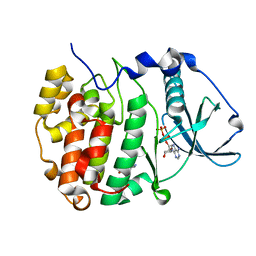 | | Crystal structure of compound 7 bound to CK2alpha | | Descriptor: | 2-(5-bromanyl-1~{H}-indol-3-yl)ethanenitrile, ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha | | Authors: | Brear, P, Fusco, C, Atkinson, E, Rossmann, M, Francis, N, Iegre, J, Hyvonen, M, Spring, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
8QZE
 
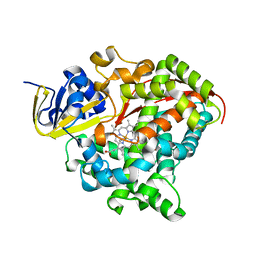 | | Heme-domain BM3 variant 21B3_F87V-A328F | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Opperman, D.J, Ebrecht, A.C, Aschenbrenner, J.C. | | Deposit date: | 2023-10-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Revisiting strategies and their combinatorial effect for introducing peroxygenase activity in CYP102A1 (P450BM3)
Mol Catal, 557, 2024
|
|
6H98
 
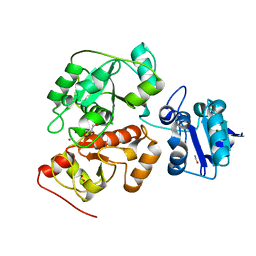 | | Native crystal structure of anaerobic ergothioneine biosynthesis enzyme from Chlorobium limicola. | | Descriptor: | CHLORIDE ION, ETHANOL, FORMIC ACID, ... | | Authors: | Leisinger, F, Burn, R, Meury, M, Lukat, P, Seebeck, F.P. | | Deposit date: | 2018-08-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Basis for Anaerobic Ergothioneine Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
7SC3
 
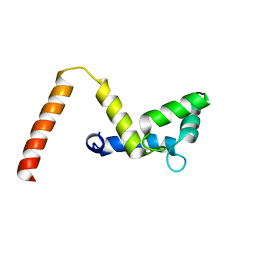 | |
7LYV
 
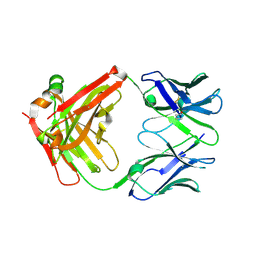 | |
6DOV
 
 | |
8RTZ
 
 | | The structure of E. coli penicillin binding protein 3 (PBP3) in complex with a bicyclic peptide inhibitor | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Bicyclic peptide inhibitor, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Rowland, C.E, Dods, R, Lewis, N, Stanway, S.J, Bellini, D, Beswick, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and chemical optimisation of a Potent, Bi-cyclic (Bicycle) Antimicrobial Inhibitor of Escherichia coli PBP3
To Be Published
|
|
6DP8
 
 | |
7SEA
 
 | | Crystal structure of human Fibrillarin in complex with compound 2 from cocktail soak | | Descriptor: | 6-(trifluoromethyl)pyrimidin-4-amine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
6DPP
 
 | |
7LHP
 
 | |
6HBB
 
 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, SULFATE ION | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
7SE9
 
 | | Crystal structure of human Fibrillarin in complex with compound 1 from single soak | | Descriptor: | (5S)-3-methyl-7-(trifluoromethyl)pyrrolo[1,2-a]pyrazin-1(2H)-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
6HBE
 
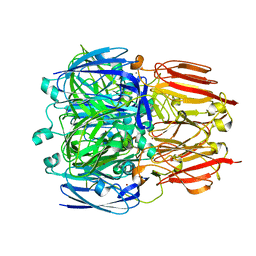 | |
6HE1
 
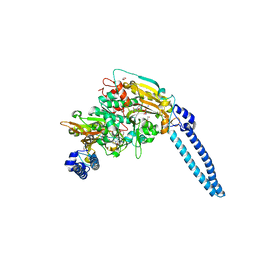 | | Pseudomonas aeruginosa Seryl-tRNA Synthetase in Complex with 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine, SODIUM ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
7LGN
 
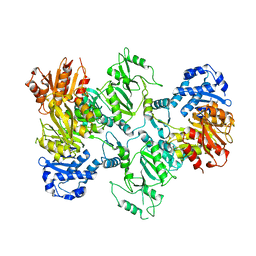 | |
7S16
 
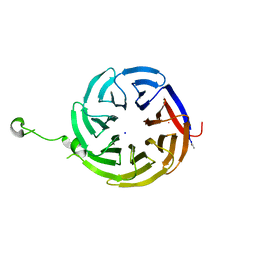 | | Crystal structure of alpha-COP-WD40 domain R57A mutant | | Descriptor: | Coatomer subunit alpha, SODIUM ION | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7ZGI
 
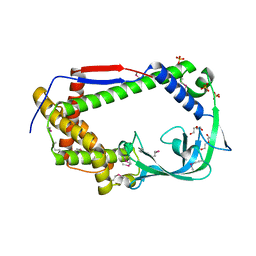 | | chloroplast trigger factor (TIG1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidylprolyl isomerase, SULFATE ION | | Authors: | Carius, Y, Ries, F, Gries, K, Trentmann, O, Willmund, F, Lancaster, C.R.D. | | Deposit date: | 2022-04-03 | | Release date: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural features of chloroplast trigger factor determined at 2.6 angstrom resolution.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
