1Q2H
 
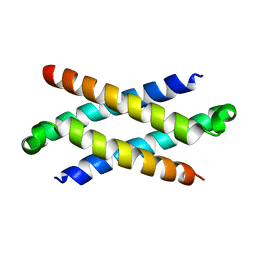 | | Phenylalanine Zipper Mediates APS Dimerization | | Descriptor: | adaptor protein with pleckstrin homology and src homology 2 domains | | Authors: | Dhe-Paganon, S, Werner, E.D, Nishi, M, Chi, Y.-I, Shoelson, S.E. | | Deposit date: | 2003-07-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A phenylalanine zipper mediates APS dimerization.
Nat.Struct.Mol.Biol., 11, 2004
|
|
8SPJ
 
 | |
2OAI
 
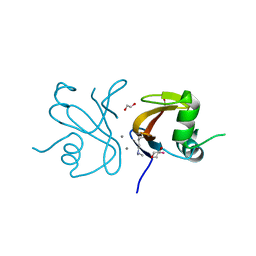 | | The structure of transporter associated domain CorC_HlyC from a Xylella fastidiosa Temecula1 hemolysin. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Hemolysin | | Authors: | Cuff, M.E, Volkart, L, Abdullah, J, Binkowski, T.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-15 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of transporter associated domain CorC_HlyC from a Xylella fastidiosa Temecula1 hemolysin.
TO BE PUBLISHED
|
|
2J4F
 
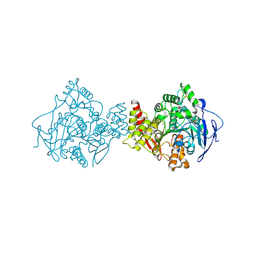 | | Torpedo acetylcholinesterase - Hg heavy-atom derivative | | Descriptor: | ACETYLCHOLINESTERASE, MERCURY (II) ION | | Authors: | Kreimer, D.I, Dolginova, E.A, Raves, M, Sussman, J.L, Silman, I, Weiner, L. | | Deposit date: | 2006-08-30 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Metastable State of Torpedo Californica Acetylcholinesterase Generated by Modification with Organomercurials
Biochemistry, 33, 1994
|
|
2O9Z
 
 | | Crystal Structure of RebH, a FAD-dependent halogenase from Lechevalieria aerocolonigenes, the Apo form | | Descriptor: | PHOSPHATE ION, Tryptophan halogenase | | Authors: | Bitto, E, Bingman, C.A, Phillips Jr, G.N. | | Deposit date: | 2006-12-14 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | The structure of flavin-dependent tryptophan 7-halogenase RebH.
Proteins, 70, 2008
|
|
5EHE
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2015-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Intrinsic Thermodynamics and Structures of 2,4- and 3,4-Substituted Fluorinated Benzenesulfonamides Binding to Carbonic Anhydrases.
ChemMedChem, 12, 2017
|
|
2WDA
 
 | | The X-ray structure of the Streptomyces coelicolor A3 Chondroitin AC Lyase in Complex with Chondroitin sulphate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Elmabrouk, Z.H, Taylor, E.J, Vincent, F, Smith, N.L, Turkenburg, J.P, Davies, G.J, Black, G.W. | | Deposit date: | 2009-03-23 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Family 8 Polysaccharide Lyase Reveal Open and Highly Occluded Substrate-Binding Cleft Conformations.
Proteins, 79, 2011
|
|
2CDE
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | Descriptor: | INKT-TCR | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
3CEV
 
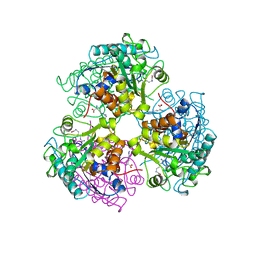 | | ARGINASE FROM BACILLUS CALDEVELOX, COMPLEXED WITH L-ARGININE | | Descriptor: | ARGININE, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
1MNO
 
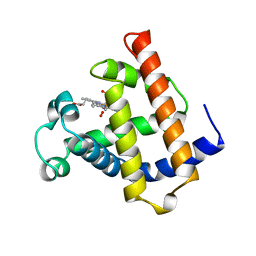 | | V68N MYOGLOBIN OXY FORM | | Descriptor: | OXYGEN MOLECULE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
7L08
 
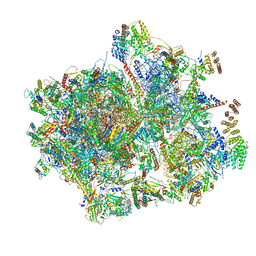 | | Cryo-EM structure of the human 55S mitoribosome-RRFmt complex. | | Descriptor: | 12S rRNA, 16S RRNA, 28S ribosomal protein S10, ... | | Authors: | Koripella, R, Agrawal, E.K, Deep, A, Agrawal, R.K. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Distinct mechanisms of the human mitoribosome recycling and antibiotic resistance.
Nat Commun, 12, 2021
|
|
1K53
 
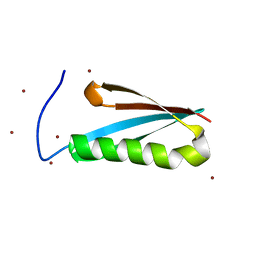 | | Monomeric Protein L B1 Domain with a G15A Mutation | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
3KOU
 
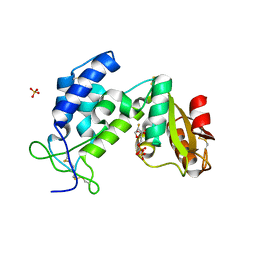 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD38 molecule, ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N.J, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-11-13 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
2WF8
 
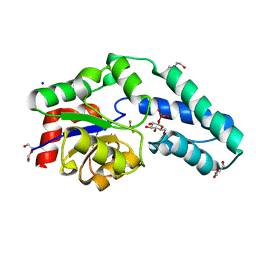 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, Glucose-1-phosphate and Beryllium trifluoride | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Near attack conformers dominate beta-phosphoglucomutase complexes where geometry and charge distribution reflect those of substrate.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
2VP0
 
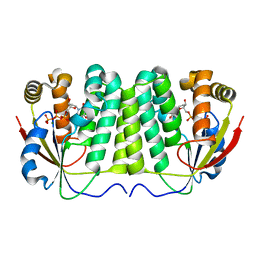 | |
7L1K
 
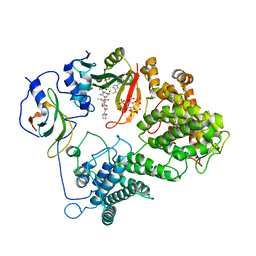 | |
4AA2
 
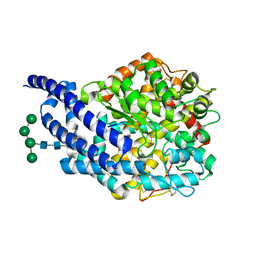 | | Crystal structure of ANCE in complex with bradykinin potentiating peptide b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, BRADYKININ-POTENTIATING PEPTIDE B, ... | | Authors: | Isaac, R.E, Akif, M, Schwager, S.L.U, Masuyer, G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2011-11-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of Peptide Recognition by the Angiotensin-I Converting Enzyme Homologue Ance from Drosophila Melanogaster
FEBS J., 279, 2012
|
|
2C8D
 
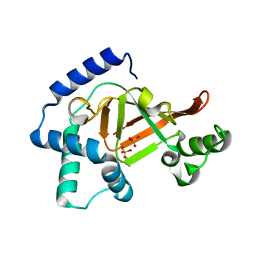 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
5SB6
 
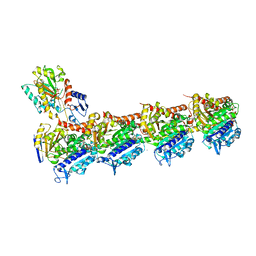 | | Tubulin-todalam-10-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muehlethaler, T, Milanos, L, Ortega, J.A, Blum, T.B, Gioia, D, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2021-07-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5SB3
 
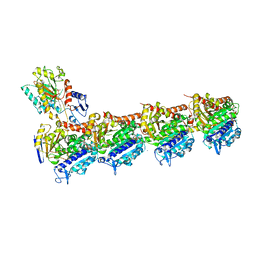 | | Tubulin-todalam-4-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muehlethaler, T, Milanos, L, Ortega, J.A, Blum, T.B, Gioia, D, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2021-07-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5SB5
 
 | | Tubulin-todalam-9-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muehlethaler, T, Milanos, L, Ortega, J.A, Blum, T.B, Gioia, D, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2021-07-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5SB4
 
 | | Tubulin-todalam-8-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muehlethaler, T, Milanos, L, Ortega, J.A, Blum, T.B, Gioia, D, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2021-07-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5ZF7
 
 | | Structure of human dihydroorotate dehydrogenase in complex with 277-9-OH | | Descriptor: | 3-chloro-4,6-dihydroxy-5-[(2E,6E,8S)-8-hydroxy-3,7-dimethylnona-2,6-dien-1-yl]-2-methylbenzaldehyde, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Miyazaki, Y, Inaoka, K.D, Shiba, T, Saimoto, H, Amalia, E, Kido, Y, Sakai, C, Nakamura, M, Moore, L.A, Harada, S, Kita, K. | | Deposit date: | 2018-03-02 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Selective Cytotoxicity of Dihydroorotate Dehydrogenase Inhibitors to Human Cancer Cells Under Hypoxia and Nutrient-Deprived Conditions.
Front Pharmacol, 9, 2018
|
|
8SXO
 
 | |
2J9J
 
 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
