4J93
 
 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BI-1 | | Descriptor: | (4S)-3-phenyl-4-(pyridin-3-yl)-4,5-dihydropyrrolo[3,4-c]pyrazol-6(2H)-one, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein | | Authors: | Lemke, C.T. | | Deposit date: | 2013-02-15 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Novel Small-Molecule HIV-1 Replication Inhibitors That Stabilize Capsid Complexes.
Antimicrob.Agents Chemother., 57, 2013
|
|
3R02
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 7-[(cis-4-aminocyclohexyl)amino]-5-bromo-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R4B
 
 | | Crystal Structure of Wild-type HIV-1 Protease in Complex With TMC310911 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({2-[(1-cyclopentylpiperidin-4-yl)amino]-1,3-benzothiazol-6-yl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-p henylbutan-2-yl}carbamate, HIV-1 protease, PHOSPHATE ION | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-03-17 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TMC310911, a Novel Human Immunodeficiency Virus Type 1 Protease Inhibitor, Shows In Vitro an Improved Resistance Profile and Higher Genetic Barrier to Resistance Compared with Current Protease Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
3NU9
 
 | |
1W2C
 
 | | Human Inositol (1,4,5) trisphosphate 3-kinase complexed with Mn2+/AMPPNP/Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, MANGANESE (II) ION, ... | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
3O9D
 
 | | Crystal Structure of wild-type HIV-1 Protease in complex with kd19 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
1RV7
 
 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-12 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
1SBG
 
 | |
2F6W
 
 | | Protein tyrosine phosphatase 1B with sulfamic acid inhibitors | | Descriptor: | (2-METHYL-5-PHENYL-2H-PYRAZOL-3-YL)-SULFAMIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Klopfenstein, S.R. | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 1,2,3,4-Tetrahydroisoquinolinyl sulfamic acids as phosphatase PTP1B inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1TV6
 
 | | HIV-1 Reverse Transcriptase Complexed with CP-94,707 | | Descriptor: | 3-[4-(2-METHYL-IMIDAZO[4,5-C]PYRIDIN-1-YL)BENZYL]-3H-BENZOTHIAZOL-2-ONE, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit | | Authors: | Pata, J.D, Stirtan, W.G, Goldstein, S.W, Steitz, T.A. | | Deposit date: | 2004-06-28 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant RTs resistant to other non-nucleoside inhibitors
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2POV
 
 | | The crystal structure of the human carbonic anhydrase II in complex with 4-amino-6-chloro-benzene-1,3-disulfonamide | | Descriptor: | 4-AMINO-6-CHLOROBENZENE-1,3-DISULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2007-04-27 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Carbonic anhydrase inhibitors: Inhibition of human, bacterial, and archaeal isozymes with benzene-1,3-disulfonamides-Solution and crystallographic studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PY6
 
 | |
2Q2Y
 
 | | Crystal Structure of KSP in complex with Inhibitor 1 | | Descriptor: | 1-{(3R,3AR)-3-[3-(4-ACETYLPIPERAZIN-1-YL)PROPYL]-8-FLUORO-3-PHENYL-3A,4-DIHYDRO-3H-PYRAZOLO[5,1-C][1,4]BENZOXAZIN-2-YL}ETHANONE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2007-05-29 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 8: Design and synthesis of 1,4-diaryl-4,5-dihydropyrazoles as potent inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3QIN
 
 | |
2QI4
 
 | | Crystal structure of protease inhibitor, MIT-2-AD93 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-3-HYDROXYBENZAMIDE, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
1WYX
 
 | | The Crystal Structure of the p130Cas SH3 Domain at 1.1 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CRK-associated substrate, MAGNESIUM ION | | Authors: | Wisniewska, M, Bossenmaier, B, Georges, G, Hesse, F, Dangl, M, Kuenkele, K.P, Ioannidis, I, Huber, R, Engh, R.A. | | Deposit date: | 2005-02-17 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The 1.1 A resolution crystal structure of the p130cas SH3 domain and ramifications for ligand selectivity
J.Mol.Biol., 347, 2005
|
|
3B3A
 
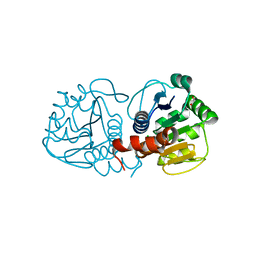 | | Structure of E163K/R145E DJ-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein DJ-1 | | Authors: | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | Deposit date: | 2007-10-19 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
4BHG
 
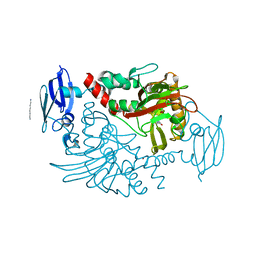 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 3-(1-Ethyl-1,1-dimethylhydrazin-1-ium-2-yl)propanoate | | Descriptor: | 3-(1-Ethyl-1,1-dimethylhydrazin-1-ium-2-yl)propanoic acid, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three Dimensional Structure of Human Gamma-Butyrobetaine Hydroxylase in Complex with 3-(1-Ethyl-1,1-Dimethylhydrazin-1-Ium-2-Yl)Propanoate
To be Published
|
|
1RD4
 
 | | An allosteric inhibitor of LFA-1 bound to its I-domain | | Descriptor: | 1-ACETYL-4-(4-{4-[(2-ETHOXYPHENYL)THIO]-3-NITROPHENYL}PYRIDIN-2-YL)PIPERAZINE, Integrin alpha-L | | Authors: | Crump, M.P, Ceska, T.A, Spyracopoulos, L, Henry, A, Archibald, S.C, Alexander, R, Taylor, R.J, Findlow, S.C, O'Connell, J, Robinson, M.K, Shock, A. | | Deposit date: | 2003-11-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an allosteric inhibitor of LFA-1 bound to the I-domain studied by crystallography, NMR, and calorimetry
Biochemistry, 43, 2004
|
|
1UZH
 
 | | A CHIMERIC CHLAMYDOMONAS, SYNECHOCOCCUS RUBISCO ENZYME | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2004-03-12 | | Release date: | 2005-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chimeric Small Subunits Influence Catalysis without Causing Global Conformational Changes in the Crystal Structure of Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase
Biochemistry, 44, 2005
|
|
3NGB
 
 | |
3RDP
 
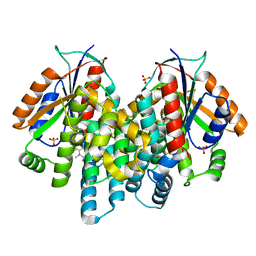 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with N-METHYL-FHBT | | Descriptor: | 6-[(2R)-2-(fluoromethyl)-3-hydroxy-propyl]-1,5-dimethyl-pyrimidine-2,4-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Perozzo, R, Westermaier, Y, Martic, M, Ametamey, S, Scapozza, L. | | Deposit date: | 2011-04-01 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, crystal structure, and in vitro biological evaluation of C-6 pyrimidine derivatives: new lead structures for monitoring gene expression in vivo.
Nucleosides Nucleotides Nucleic Acids, 30, 2011
|
|
2O4P
 
 | | Crystal Structure of HIV-1 Protease (Q7K) in Complex with Tipranavir | | Descriptor: | GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, protease | | Authors: | Kang, L.W, Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
1PRO
 
 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-98881 | | Descriptor: | (5R,6R)-2,4-BIS-(4-HYDROXY-3-METHOXYBENZYL)-1,5-DIBENZYL-3-OXO-6-HYDROXY-1,2,4-TRIAZACYCLOHEPTANE, HIV-1 PROTEASE | | Authors: | Park, C.H, Kong, X.P, Dealwis, C.G. | | Deposit date: | 1995-07-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel, picomolar inhibitor of human immunodeficiency virus type 1 protease.
J.Med.Chem., 39, 1996
|
|
3R04
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-{6-[(trans-4-aminocyclohexyl)amino]pyrazin-2-yl}-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
