1C6Z
 
 | | ALTERNATE BINDING SITE FOR THE P1-P3 GROUP OF A CLASS OF POTENT HIV-1 PROTEASE INHIBITORS AS A RESULT OF CONCERTED STRUCTURAL CHANGE IN 80'S LOOP. | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEIN (PROTEASE) | | Authors: | Munshi, S. | | Deposit date: | 1999-12-28 | | Release date: | 2000-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alternate binding site for the P1-P3 group of a class of potent HIV-1 protease inhibitors as a result of concerted structural change in the 80s loop of the protease.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C9W
 
 | | CHO REDUCTASE WITH NADP+ | | Descriptor: | CHO REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Li, X, Hyndman, D, Flynn, T.G, Jia, Z. | | Deposit date: | 1999-08-03 | | Release date: | 2000-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
Proteins, 38, 2000
|
|
1CAG
 
 | | CRYSTAL AND MOLECULAR STRUCTURE OF A COLLAGEN-LIKE PEPTIDE AT 1.9 ANGSTROM RESOLUTION | | Descriptor: | ACETIC ACID, COLLAGEN-LIKE PEPTIDE | | Authors: | Bella, J, Eaton, M, Brodsky, B, Berman, H.M. | | Deposit date: | 1994-03-29 | | Release date: | 1994-11-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution.
Science, 266, 1994
|
|
1C6Y
 
 | |
1C94
 
 | | REVERSING THE SEQUENCE OF THE GCN4 LEUCINE ZIPPER DOES NOT AFFECT ITS FOLD. | | Descriptor: | RETRO-GCN4 LEUCINE ZIPPER | | Authors: | Mittl, P.R.E, Deillon, C.A, Sargent, D, Liu, N, Klauser, S, Thomas, R.M, Gutte, B, Gruetter, M.G. | | Deposit date: | 1999-07-30 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C97
 
 | | S642A:ISOCITRATE COMPLEX OF ACONITASE | | Descriptor: | IRON/SULFUR CLUSTER, ISOCITRIC ACID, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1CA7
 
 | |
1AC5
 
 | | CRYSTAL STRUCTURE OF KEX1(DELTA)P, A PROHORMONE-PROCESSING CARBOXYPEPTIDASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, KEX1(DELTA)P | | Authors: | Shilton, B.H, Thomas, D.Y, Cygler, M. | | Deposit date: | 1997-02-13 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Kex1deltap, a prohormone-processing carboxypeptidase from Saccharomyces cerevisiae.
Biochemistry, 36, 1997
|
|
1AJH
 
 | | PHOTOPRODUCT OF CARBONMONOXY MYOGLOBIN AT 40 K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teng, T.Y, Srajer, V, Moffat, K. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Photolysis-induced structural changes in single crystals of carbonmonoxy myoglobin at 40 K.
Nat.Struct.Biol., 1, 1994
|
|
1AD0
 
 | | FAB FRAGMENT OF ENGINEERED HUMAN MONOCLONAL ANTIBODY A5B7 | | Descriptor: | ANTIBODY A5B7 (HEAVY CHAIN), ANTIBODY A5B7 (LIGHT CHAIN) | | Authors: | Banfield, M.J, Brady, R.L. | | Deposit date: | 1997-02-19 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs.
Proteins, 29, 1997
|
|
1A02
 
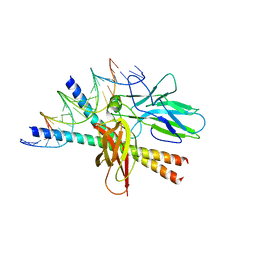 | | STRUCTURE OF THE DNA BINDING DOMAINS OF NFAT, FOS AND JUN BOUND TO DNA | | Descriptor: | AP-1 FRAGMENT FOS, AP-1 FRAGMENT JUN, DNA (5'-D(*DAP*DAP*DCP*DTP*DAP*DTP*DGP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DCP*DC)-3'), ... | | Authors: | Chen, L, Glover, J.N.M, Hogan, P.G, Rao, A, Harrison, S.C. | | Deposit date: | 1997-12-08 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA.
Nature, 392, 1998
|
|
1A1C
 
 | |
1AFA
 
 | |
1ALW
 
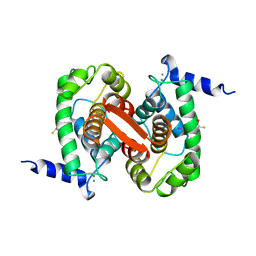 | | INHIBITOR AND CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | Descriptor: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, CALPAIN | | Authors: | Narayana, S.V.L, Lin, G. | | Deposit date: | 1997-06-04 | | Release date: | 1998-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
1A1K
 
 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GACC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*CP*CP*AP*CP*GP*C)-3'), RADR ZIF268 VARIANT, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1V
 
 | | HEPATITIS C VIRUS NS3 HELICASE DOMAIN COMPLEXED WITH SINGLE STRANDED SDNA | | Descriptor: | DNA (5'-D(*UP*UP*UP*UP*UP*UP*UP*U)-3'), PROTEIN (NS3 PROTEIN), SULFATE ION | | Authors: | Kim, J.L, Morgenstern, K.A, Griffith, J.P, Dwyer, M.D, Thomson, J.A, Murcko, M.A, Lin, C, Caron, P.R. | | Deposit date: | 1997-12-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding.
Structure, 6, 1998
|
|
1A3R
 
 | |
1A43
 
 | | STRUCTURE OF THE HIV-1 CAPSID PROTEIN DIMERIZATION DOMAIN AT 2.6A RESOLUTION | | Descriptor: | HIV-1 CAPSID | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-02-10 | | Release date: | 1999-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1A4Z
 
 | |
1AIB
 
 | |
1A81
 
 | |
1A94
 
 | | STRUCTURAL BASIS FOR SPECIFICITY OF RETROVIRAL PROTEASES | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE | | Authors: | Wu, J, Adomat, J.M, Ridky, T.W, Louis, J.M, Leis, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 1998-04-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for specificity of retroviral proteases.
Biochemistry, 37, 1998
|
|
15C8
 
 | | CATALYTIC ANTIBODY 5C8, FREE FAB | | Descriptor: | IGG 5C8 FAB (HEAVY CHAIN), IGG 5C8 FAB (LIGHT CHAIN) | | Authors: | Gruber, K, Wilson, I.A. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-Induced Conformational Changes in a Catalytic Antibody: Comparison of the Bound and Unbound Structure of Fab 5C8
To be Published
|
|
1ANN
 
 | | ANNEXIN IV | | Descriptor: | ANNEXIN IV, CALCIUM ION | | Authors: | Sutton, R.B, Sprang, S.R. | | Deposit date: | 1995-09-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three Dimensional Structure of Annexin IV
Annexins: Molecular Structure to Cellular Function, 1996
|
|
1ADU
 
 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
