7A71
 
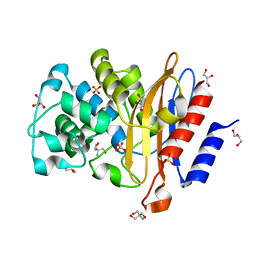 | | Structure of G132S BlaC from Mycobacterium tuberculosis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The G132S Mutation Enhances the Resistance of Mycobacterium tuberculosis beta-Lactamase against Sulbactam.
Biochemistry, 60, 2021
|
|
8DVI
 
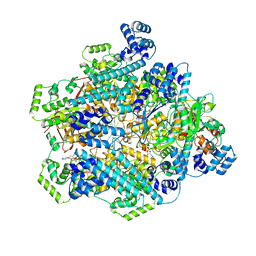 | | T4 bacteriophage primosome with single strand DNA, State 2 | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (70-MER), DNA primase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-29 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
7MCS
 
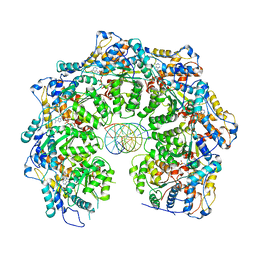 | | Cryo-electron microscopy structure of TnsC(1-503)A225V bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Shen, Y, Ortega, J, Guarne, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-02-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for DNA targeting by the Tn7 transposon.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6LTB
 
 | |
7MBW
 
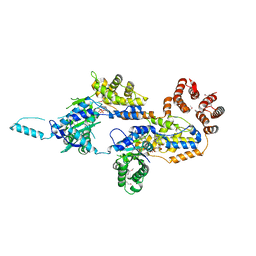 | | Crystal structure of TnsC(1-503)A225V | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transposon Tn7 transposition protein TnsC | | Authors: | Shen, Y, Guarne, A. | | Deposit date: | 2021-04-01 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-02 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for DNA targeting by the Tn7 transposon.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6LTA
 
 | |
6UT9
 
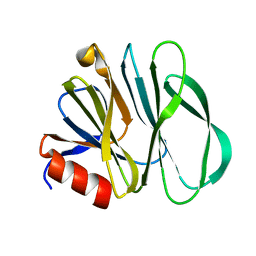 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[4] rotavirus strain BM5265 | | Descriptor: | Outer capsid protein VP4 | | Authors: | Xu, S, Stuckert, M, Burnside, R, McGinnis, K, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6PHE
 
 | |
8DVC
 
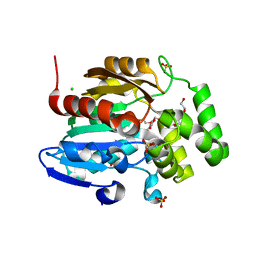 | | Receptor ShHTL5 from Striga hermonthica in complex with strigolactone agonist GR24 | | Descriptor: | (3R,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methyl)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Arellano-Saab, A, Skarina, T, Yim, V, Savchenko, A, Stogios, P.J, McCourt, P. | | Deposit date: | 2022-07-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.638 Å) | | Cite: | Structural analysis of a hormone-bound Striga strigolactone receptor.
Nat.Plants, 9, 2023
|
|
7A3K
 
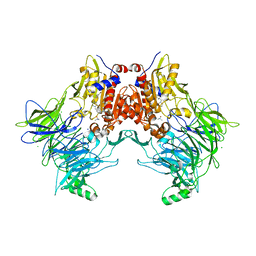 | |
7MHU
 
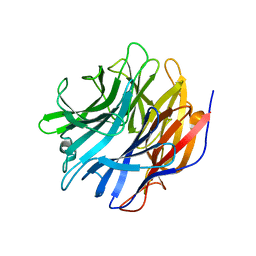 | | Sialidase24 apo | | Descriptor: | Exo-alpha-sialidase | | Authors: | Rees, S.D, Chang, G.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational models in the service of X-ray and cryo-electron microscopy structure determination.
Proteins, 89, 2021
|
|
6LZZ
 
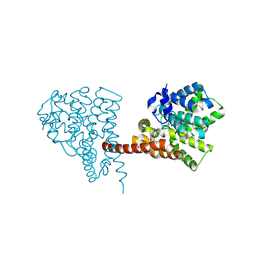 | | Crystal structure of the PDE9 catalytic domain in complex with inhibitor 4a | | Descriptor: | 1-cyclopentyl-6-[[(2R)-1-(2-oxa-6-azaspiro[3.3]heptan-6-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Huang, Y.Y, Wu, Y, Luo, H.B. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.40003753 Å) | | Cite: | Identification of phosphodiesterase-9 as a novel target for pulmonary arterial hypertension by using highly selective and orally bioavailable inhibitors
To Be Published
|
|
8DVF
 
 | | T4 Bacteriophage primosome with single strand DNA, state 1 | | Descriptor: | DNA (5'-D(P*GP*GP*CP*TP*G)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA primase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
6VDR
 
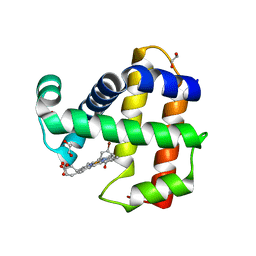 | | Crystal Structure of Dehaloperoxidase B in Complex with cofactor Iron(III) Deuteroporphyrin IX and Substrate 4-bromophenol | | Descriptor: | 1,2-ETHANEDIOL, 4-BROMOPHENOL, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, McGuire, A, Malewschik, T. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Nonnative Heme Incorporation into Multifunctional Globin Increases Peroxygenase Activity an Order and Magnitude Compared to Native Enzyme
To Be Published
|
|
6P8V
 
 | | Structure of E. coli MS115-1 HORMA:CdnC:Trip13 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, AAA family, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6VE1
 
 | |
6M38
 
 | | X-ray structure of a Drosophila dopamine transporter with subsiteB mutations (D121G/S426M) in S-duloxetine bound form | | Descriptor: | (3S)-N-methyl-3-(naphthalen-1-yloxy)-3-(thiophen-2-yl)propan-1-amine, Antibody fragment 9D5 heavy chain, Antibody fragment 9D5 light chain, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-03-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
7MD4
 
 | | Insulin receptor ectodomain dimer complexed with two IRPA-3 partial agonists | | Descriptor: | Insulin B chain, Insulin chain A, Isoform Short of Insulin receptor, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
6ZZC
 
 | | MB_CRS6-1 bound to CrSAS-6_6HR | | Descriptor: | Centriole protein, DODECAETHYLENE GLYCOL, MB_CrS6-1 | | Authors: | Hatzopoulos, G.N, Kukenshoner, T, Banterle, N, Favez, T, Fluckiger, I, Hantschel, O, Gonczy, P. | | Deposit date: | 2020-08-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Nat Commun, 12, 2021
|
|
8E4V
 
 | | Solution structure of the WH domain of MORF | | Descriptor: | Isoform 3 of Histone acetyltransferase KAT6B | | Authors: | Zhang, Y, Kutateladze, T.G. | | Deposit date: | 2022-08-19 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | MORF and MOZ acetyltransferases target unmethylated CpG islands through the winged helix domain.
Nat Commun, 14, 2023
|
|
6P9U
 
 | | Crystal structure of human thrombin mutant W215A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, ZINC ION | | Authors: | Pelc, L.A, Koester, S.K, Chen, Z, Di Cera, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Residues W215, E217 and E192 control the allosteric E*-E equilibrium of thrombin.
Sci Rep, 9, 2019
|
|
6VEP
 
 | | Human insulin in complex with the human insulin microreceptor in turn in complex with Fv 83-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lawrence, M.C, Menting, J.G.T. | | Deposit date: | 2020-01-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structurally minimized yet fully active insulin based on cone-snail venom insulin principles.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6M3Z
 
 | | X-ray structure of a Drosophila dopamine transporter with NET-like mutations (D121G/S426M/F471L) in milnacipran bound form | | Descriptor: | (1R,2S)-2-(aminomethyl)-N,N-diethyl-1-phenyl-cyclopropane-1-carboxamide, Antibody fragment 9D5 Light chain, Antibody fragment 9D5 heavy chain, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-03-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
6PIJ
 
 | | Target DNA-bound V. cholerae TniQ-Cascade complex, closed conformation | | Descriptor: | Non-targeting strand ssDNA, Targeting strand ssDNA, TniQ monomer 2, ... | | Authors: | Halpin-Healy, T, Klompe, S, Sternberg, S.H. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system.
Nature, 577, 2020
|
|
8DTP
 
 | | Close state of T4 bacteriophage gp41 hexamer bound with single strand DNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DnaB-like replicative helicase, MAGNESIUM ION, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
