5F5C
 
 | | Crystal Structure of human JMJD2D complexed with KDOPP7 | | Descriptor: | 1,2-ETHANEDIOL, 8-[[(phenylmethyl)amino]methyl]-1~{H}-pyrido[3,4-d]pyrimidin-4-one, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
2W7N
 
 | | Crystal Structure of KorA Bound to Operator DNA: Insight into Repressor Cooperation in RP4 Gene Regulation | | Descriptor: | 5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*GP*CP *TP*AP*AP*AP*CP*AP*AP*G)-3', 5'-D(*CP*BRUP*BRUP*GP*TP*TP*TP*AP*GP*CP*TP*AP *AP*AP*CP*AP*BRUP*T)-3', TRFB TRANSCRIPTIONAL REPRESSOR PROTEIN | | Authors: | Koenig, B, Mueller, J.J, Lanka, E, Heinemann, U. | | Deposit date: | 2008-12-23 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Kora Bound to Operator DNA: Insight Into Repressor Cooperation in Rp4 Gene Regulation
Nucleic Acids Res., 37, 2009
|
|
5XJG
 
 | | Crystal structure of Vac8p bound to Nvj1p | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Nucleus-vacuole junction protein 1, ... | | Authors: | Jeong, H, Park, J, Jun, Y, Lee, C. | | Deposit date: | 2017-05-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4Q8N
 
 | |
7NVO
 
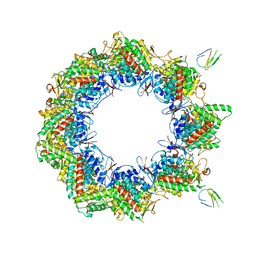 | | Human TRiC complex in open state with nanobody bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4GJJ
 
 | |
6WPK
 
 | |
2ZMX
 
 | |
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7NVM
 
 | | Human TRiC complex in closed state with nanobody Nb18, actin and PhLP2A bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Actin, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6WL0
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3-RD | | Descriptor: | peptide 36-31-3-RD | | Authors: | Wang, F, Gnewou, O.M, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
3FBM
 
 | | D431N Mutant VP2 Protein of Infectious Bursal Disease Virus; Derived T=1 Particles | | Descriptor: | CALCIUM ION, Polyprotein | | Authors: | Irigoyen, N, Garriga, D, Navarro, A, Verdaguer, N, Rodriguez, J.F, Caston, J.R. | | Deposit date: | 2008-11-19 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Autoproteolytic Activity Derived from the Infectious Bursal Disease Virus Capsid Protein
J.Biol.Chem., 284, 2009
|
|
5EZ3
 
 | |
5XUB
 
 | |
2NB1
 
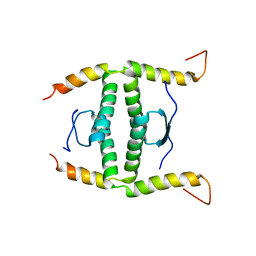 | | P63/p73 hetero-tetramerisation domain | | Descriptor: | Tumor protein 63, Tumor protein p73 | | Authors: | Gebel, J, Buchner, L, Loehr, F.M, Luh, L.M, Coutandin, D, Guentert, P, Doetsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
5F0J
 
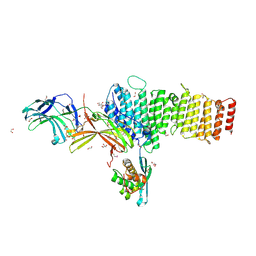 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
4PY8
 
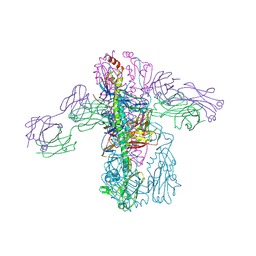 | | Crystal structure of Fab 3.1 in complex with the 1918 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Dreyfus, C. | | Deposit date: | 2014-03-26 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Alternative Recognition of the Conserved Stem Epitope in Influenza A Virus Hemagglutinin by a VH3-30-Encoded Heterosubtypic Antibody.
J.Virol., 88, 2014
|
|
1PQX
 
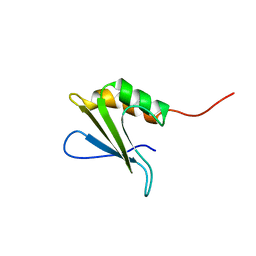 | | Solution NMR Structure of Staphylococcus aureus protein SAV1430. Northeast Structural Genomics Consortium Target ZR18. | | Descriptor: | conserved hypothetical protein | | Authors: | Baran, M.C, Aramini, J.M, Xiao, R, Huang, Y.J, Acton, T.B, Shih, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-06-19 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutre of the Hypothetical Staphylococcus Aureus protein SAV1430. Northest Strucutral Genomics Consortium target ZR18
To be Published
|
|
4XLF
 
 | | Tailspike protein double mutant D339N/E372A of E. coli bacteriophage HK620 in complex with pentasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-13 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
3FC3
 
 | | Crystal structure of the beta-beta-alpha-Me type II restriction endonuclease Hpy99I | | Descriptor: | 5'-(*DCP*DTP*DCP*DGP*DAP*DCP*DGP*DTP*DAP*DGP*DA)-3', 5'-(*DTP*DAP*DCP*DGP*DTP*DCP*DGP*DAP*DGP*DTP*DC)-3', Restriction endonuclease Hpy99I, ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA.
Nucleic Acids Res., 37, 2009
|
|
4XLZ
 
 | | N,N'-diacetylchitobiose deacetylase (SeMet derivative) from Pyrococcus furiosus in the presence of cadmium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7NSG
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | | Descriptor: | (+)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, (-)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S.M.M, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
TO BE PUBLISHED
|
|
5F78
 
 | | Crystal structure of Mutant N87T of adenosine/Methylthioadenosine phosphorylase from Schistosoma mansoni in APO form | | Descriptor: | Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8518 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
4Y2X
 
 | | Structure of soluble epoxide hydrolase in complex with 2-({[2-(adamantan-1-yl)ethyl]amino}methyl)phenol | | Descriptor: | 2-[({2-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-yl]ethyl}amino)methyl]phenol, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of N-ethylmethylamine as a novel scaffold for inhibitors of soluble epoxide hydrolase by crystallographic fragment screening
Bioorg.Med.Chem., 23, 2015
|
|
3FCU
 
 | | Structure of headpiece of integrin aIIBb3 in open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
