1HVN
 
 | |
1HVO
 
 | |
1O7Q
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | Deposit date: | 2002-11-12 | | Release date: | 2003-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
1WYX
 
 | | The Crystal Structure of the p130Cas SH3 Domain at 1.1 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CRK-associated substrate, MAGNESIUM ION | | Authors: | Wisniewska, M, Bossenmaier, B, Georges, G, Hesse, F, Dangl, M, Kuenkele, K.P, Ioannidis, I, Huber, R, Engh, R.A. | | Deposit date: | 2005-02-17 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The 1.1 A resolution crystal structure of the p130cas SH3 domain and ramifications for ligand selectivity
J.Mol.Biol., 347, 2005
|
|
1EFN
 
 | |
1RXO
 
 | | ACTIVATED SPINACH RUBISCO IN COMPLEX WITH ITS SUBSTRATE RIBULOSE-1,5-BISPHOSPHATE AND CALCIUM | | Descriptor: | CALCIUM ION, RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE, RIBULOSE-1,5-DIPHOSPHATE | | Authors: | Taylor, T.C, Andersson, I. | | Deposit date: | 1996-12-06 | | Release date: | 1997-06-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the complex between rubisco and its natural substrate ribulose 1,5-bisphosphate.
J.Mol.Biol., 265, 1997
|
|
2AID
 
 | | STRUCTURE OF A NON-PEPTIDE INHIBITOR COMPLEXED WITH HIV-1 PROTEASE: DEVELOPING A CYCLE OF STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 4-(4-CHLORO-PHENYL)-1-{3-[2-(4-FLUORO-PHENYL)-[1,3]DITHIOLAN-2-YL]-PROPYL}-PIPERIDIN-4-OL, CHLORIDE ION, HUMAN IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Rutenber, E.E, Fauman, E.B, Keenan, R.J, Stroud, R.M. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design.
J.Biol.Chem., 268, 1993
|
|
3T1A
 
 | |
1G0R
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). THYMIDINE/GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1TW5
 
 | | beta1,4-galactosyltransferase mutant M344H-Gal-T1 in complex with Chitobiose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramakrishnan, B, Boeggeman, E, Qasba, P.K. | | Deposit date: | 2004-06-30 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effect of the Met344His mutation on the conformational dynamics of bovine beta-1,4-galactosyltransferase: crystal structure of the Met344His mutant in complex with chitobiose
Biochemistry, 43, 2004
|
|
2JID
 
 | | Human Dipeptidyl peptidase IV in complex with 1-(3,4-Dimethoxy-phenyl) -3-m-tolyl-piperidine-4-ylamine | | Descriptor: | (3R,4S)-1-(3,4-DIMETHOXYPHENYL)-3-(3-METHYLPHENYL)PIPERIDIN-4-AMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIPEPTIDYL PEPTIDASE 4 | | Authors: | Hennig, M, Stihle, M, Luebbers, T, Thoma, R. | | Deposit date: | 2007-02-28 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 1,3-Disubstituted 4-Aminopiperidines as Useful Tools in the Optimization of the 2-Aminobenzo[A]Quinolizine Dipeptidyl Peptidase Iv Inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2J50
 
 | | Structure of Aurora-2 in complex with PHA-739358 | | Descriptor: | N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE 6, SULFATE ION | | Authors: | Cameron, A.D, Izzo, G, Storici, P, Rusconi, L, Fancelli, D, Varasi, M, Berta, D, Bindi, S, Forte, B, Severino, D, Tonani, R, Vianello, P. | | Deposit date: | 2006-09-08 | | Release date: | 2006-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 1,4,5,6-Tetrahydropyrrolo[3,4-C]Pyrazoles: Identification of a Potent Aurora Kinase Inhibitor with a Favorable Antitumor Kinase Inhibition Profile.
J.Med.Chem., 49, 2006
|
|
3OXC
 
 | | Wild Type HIV-1 Protease with Antiviral Drug Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, FORMIC ACID, Protease, ... | | Authors: | Kovalevsky, A.Y, Wang, Y.-F, Tie, Y, Weber, I.T. | | Deposit date: | 2010-09-21 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir
Proteins, 67, 2007
|
|
3OXV
 
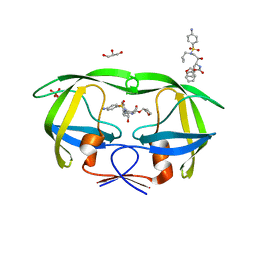 | | Crystal Structure of HIV-1 I50V, A71 Protease in Complex with the protease inhibitor amprenavir. | | Descriptor: | ACETATE ION, GLYCEROL, HIV-1 Protease, ... | | Authors: | Schiffer, C.A, Mittal, S, Bandaranayake, R.M. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
2I4U
 
 | | HIV-1 protease with TMC-126 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
3OXW
 
 | | Crystal Structure of HIV-1 I50V, A71V Protease in Complex with the Protease Inhibitor Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Mittal, S, Bandaranayake, R.M, Schiffer, C.A. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
1Q9D
 
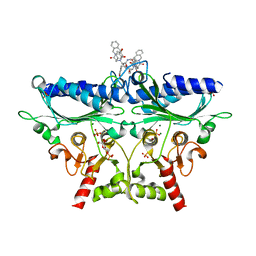 | | Fructose-1,6-bisphosphatase Complexed with a New Allosteric Site Inhibitor (I-State) | | Descriptor: | 3-(4-HYDROXYBENZYL)-2-[1-({[2-(4-HYDROXYPHENYL)ETHYL]AMINO}CARBONYL)BUTYL]-4-OXO-3,6,11,11A-TETRAHYDRO-4H-PYRAZINO[1,2-B]ISOQUINOLIN-2-IUM-1-OLATE, 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, ... | | Authors: | Honzatko, R.B, Choe, J.Y. | | Deposit date: | 2003-08-25 | | Release date: | 2003-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibition of fructose-1,6-bisphosphatase by a new class of allosteric effectors
J.Biol.Chem., 278, 2003
|
|
3V81
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and the nonnucleoside inhibitor nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8503 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1HFS
 
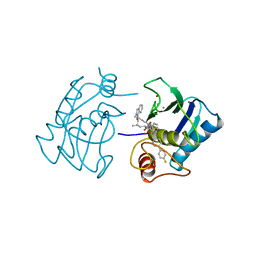 | |
3VF7
 
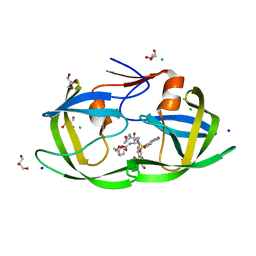 | | Crystal Structure of HIV-1 Protease Mutant L76V with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
1YPQ
 
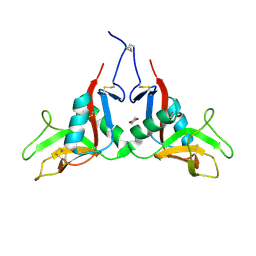 | |
3VZD
 
 | | Crystal structure of Sphingosine Kinase 1 with inhibitor and ADP | | Descriptor: | 4-{[4-(4-chlorophenyl)-1,3-thiazol-2-yl]amino}phenol, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
2UXZ
 
 | | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary- Alcohol-Containing Transition-State Mimic | | Descriptor: | HIV-1 PROTEASE, METHYL [(1S)-1-({2-[(4R)-4-BENZYL-4-HYDROXY-5-{[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]AMINO}-5-OXOPENTYL]-2-(4-BROMOBENZYL)HYDRAZINO}CARBONYL)-2,2-DIMETHYLPROPYL]CARBAMATE | | Authors: | Ginman, N, Samuelsson, B, Hallberg, A, Unge, J.T, Unge, T.K. | | Deposit date: | 2007-04-02 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary-Alcohol-Containing Transition-State Mimic.
J.Med.Chem., 51, 2008
|
|
2UY0
 
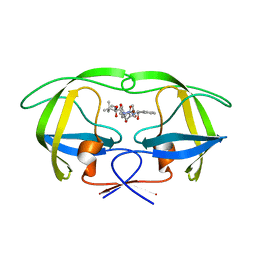 | | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary- Alcohol-Containing Transition-State Mimic | | Descriptor: | HIV-1 PROTEASE, METHYL [(1S)-1-({2-[(4R)-4-BENZYL-4-HYDROXY-5-{[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]AMINO}-5-OXOPENTANOYL]-2-(4-BROMOBENZYL)HYDRAZINO}CARBONYL)-2,2-DIMETHYLPROPYL]CARBAMATE | | Authors: | Ginman, N, Samuelsson, B, Hallberg, A, Unge, J.T, Unge, T.K. | | Deposit date: | 2007-04-02 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary-Alcohol-Containing Transition-State Mimic.
J.Med.Chem., 51, 2008
|
|
3R6B
 
 | | Crystal Structure of Thrombospondin-1 TSR Domains 2 and 3 | | Descriptor: | 1,2-ETHANEDIOL, Thrombospondin-1 | | Authors: | Page, R.C, Klenotic, P.A, Misra, S, Silverstein, R.L. | | Deposit date: | 2011-03-21 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expression, purification and structural characterization of functionally replete thrombospondin-1 type 1 repeats in a bacterial expression system.
Protein Expr.Purif., 80, 2011
|
|
