5LEM
 
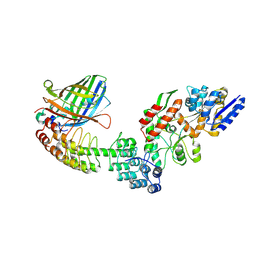 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_11_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | Descriptor: | DD_Off7_11_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
4FE8
 
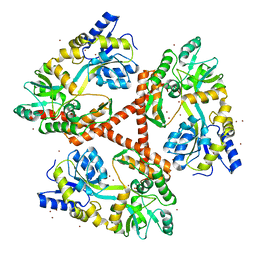 | | Crystal Structure of Htt36Q3H-EX1-X1-C1(Alpha) | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
6DBJ
 
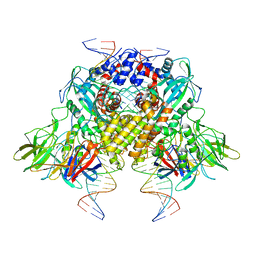 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward stand of RSS signal end, Forward strand of coding flank, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2ZXT
 
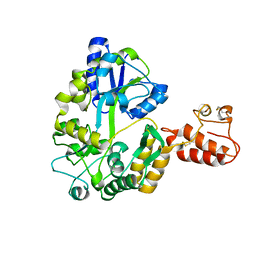 | | Crystal structure of Tim40/MIA40, a disulfide relay system in mitochondria, solved as MBP fusion protein | | Descriptor: | Maltose-binding periplasmic protein, LINKER, Mitochondrial intermembrane space import and assembly protein 40, ... | | Authors: | Kawano, S, Momose, T, Watanabe, N, Endo, T. | | Deposit date: | 2009-01-07 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5Y2G
 
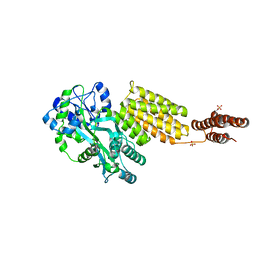 | | Structure of MBP tagged GBS CAMP | | Descriptor: | Maltose-binding periplasmic protein,Protein B, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2017-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of the CAMP factor of Streptococcus agalactiae with the aid of an MBP tag and insights into membrane-surface attachment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7W3Z
 
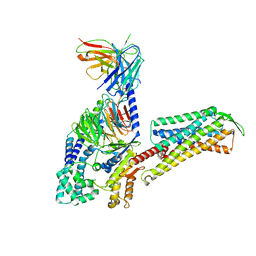 | | Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Gastrin Releasing Peptide and Gq heterotrimers | | Descriptor: | Gastrin Releasing Peptide PRGNHWAVGHLM(NH2), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhan, Y, Peng, S, Zhang, H. | | Deposit date: | 2021-11-26 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of human gastrin-releasing peptide receptors bound to antagonist and agonist for cancer and itch therapy.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7W40
 
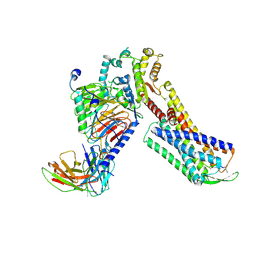 | | Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Bombesin (6-14) [D-Phe6, beta-Ala11, Phe13, Nle14] and Gq heterotrimers | | Descriptor: | Bombesin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhan, Y, Peng, S, Zhang, H. | | Deposit date: | 2021-11-26 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of human gastrin-releasing peptide receptors bound to antagonist and agonist for cancer and itch therapy.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6YSN
 
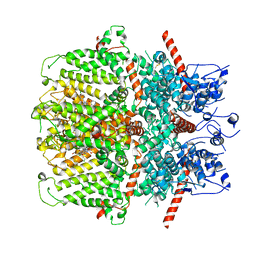 | | Human TRPC5 in complex with Pico145 (HC-608) | | Descriptor: | 7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)-8-[3-(trifluoromethyloxy)phenoxy]purine-2,6-dione, Maltose/maltodextrin-binding periplasmic protein,Short transient receptor potential channel 5 | | Authors: | Wright, D.J, Johnson, R.M, Muench, S.P, Bon, R.S. | | Deposit date: | 2020-04-22 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Human TRPC5 structures reveal interaction of a xanthine-based TRPC1/4/5 inhibitor with a conserved lipid binding site.
Commun Biol, 3, 2020
|
|
8JI0
 
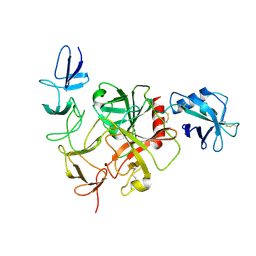 | |
4FEC
 
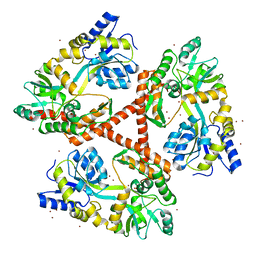 | | Crystal Structure of Htt36Q3H | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
8XB2
 
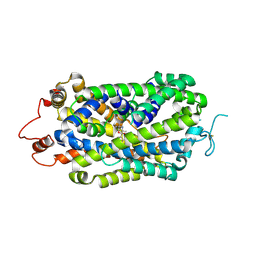 | | Structure of radafaxine-bound state of the human Norepinephrine Transporter | | Descriptor: | (2~{S},3~{S})-2-(3-chlorophenyl)-3,5,5-trimethyl-morpholin-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GFP-MBP-solute carrier family 6 member 2,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter | | Authors: | Wu, J.X, Ji, W.M. | | Deposit date: | 2023-12-05 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Substrate binding and inhibition mechanism of norepinephrine transporter
To Be Published
|
|
4BL8
 
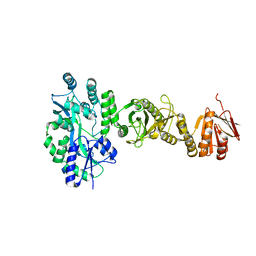 | | Crystal structure of full-length human Suppressor of fused (SUFU) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Karlstrom, M, Finta, C, Cherry, A.L, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3SEV
 
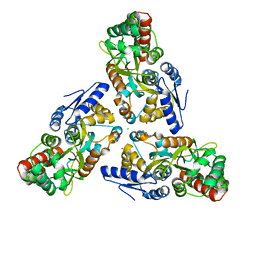 | | Zn-mediated Trimer of Maltose-binding Protein E310H/K314H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, Maltose-binding periplasmic protein, ZINC ION, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
7MN6
 
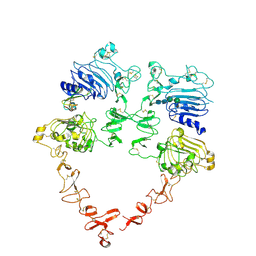 | | Structure of the HER2 S310F/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
3PV0
 
 | |
3EF7
 
 | |
5LEL
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_10_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | Descriptor: | DD_Off7_10_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
3PUY
 
 | |
6M4W
 
 | | Crystal structure of MBP fused split FKBP-FRB T2098L mutant in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
5JQE
 
 | | Crystal structure of caspase8 tDED | | Descriptor: | Sugar ABC transporter substrate-binding protein,Caspase-8 chimera | | Authors: | Fu, T, Li, Y, Lu, A, Wu, H. | | Deposit date: | 2016-05-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.157 Å) | | Cite: | Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex.
Mol. Cell, 64, 2016
|
|
5W1C
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
4KGO
 
 | |
7P1G
 
 | | Structure of the P. aeruginosa ExoY-F-actin complex | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Belyy, A, Merino, F, Raunser, S. | | Deposit date: | 2021-07-01 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of actin-dependent activation of nucleotidyl cyclase toxins from bacterial human pathogens.
Nat Commun, 12, 2021
|
|
6LF3
 
 | |
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | Descriptor: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | Authors: | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
