6QYL
 
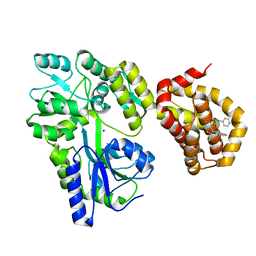 | | Structure of MBP-Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8SBU
 
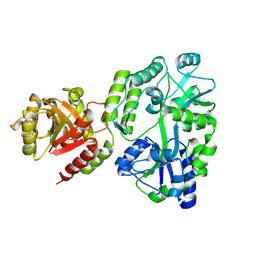 | | Crystal structure of MBP fusion with HPPK from Methanocaldococcus jannaschii | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Shaw, G.X, Needle, D, Stair, N.R, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-04-04 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of MBP fusion with HPPK from Methanocaldococcus jannaschii
To be published
|
|
3RLF
 
 | |
6WGZ
 
 | |
5IHJ
 
 | |
1E3M
 
 | | The crystal structure of E. coli MutS binding to DNA with a G:T mismatch | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP* GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP* TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Perrakis, A, Enzlin, J.H, Winterwerp, H.H.K, De Wind, N, Sixma, T.K. | | Deposit date: | 2000-06-19 | | Release date: | 2000-11-01 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of DNA Mismatch Repair Protein Muts Binding to a G X T Mismatch
Nature, 407, 2000
|
|
7CY5
 
 | | Crystal Structure of CMD1 in complex with vitamin C | | Descriptor: | ASCORBIC ACID, CITRIC ACID, FE (III) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
8H68
 
 | | Crystal structure of Caenorhabditis elegans NMAD-1 in complex with NOG and Mg(II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shi, Y, Ding, J, Yang, H. | | Deposit date: | 2022-10-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caenorhabditis elegans NMAD-1 functions as a demethylase for actin.
J Mol Cell Biol, 15, 2023
|
|
3K0S
 
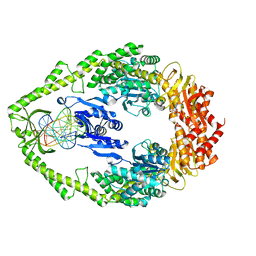 | | Crystal structure of E.coli DNA mismatch repair protein MutS, D693N mutant, in complex with GT mismatched DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*C*AP*CP*T*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Reumer, G.A, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2009-09-25 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Magnesium coordination controls the molecular switch function of DNA mismatch repair protein MutS.
J.Biol.Chem., 285, 2010
|
|
1ZKB
 
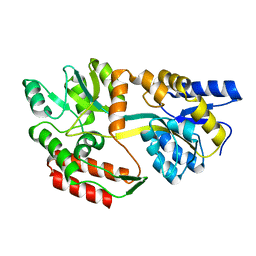 | | Zinc-free Engineered maltose binding protein | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2005-05-02 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of an engineered zinc biosensor reveal an unanticipated mode of zinc binding.
J.Mol.Biol., 354, 2005
|
|
7CY4
 
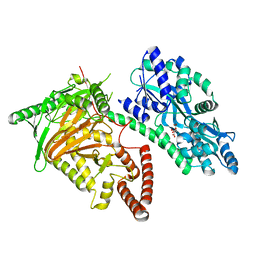 | | Crystal Structure of CMD1 in apo form | | Descriptor: | CITRIC ACID, FE (III) ION, Maltodextrin-binding protein,5-methylcytosine-modifying enzyme 1 | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
6QZ7
 
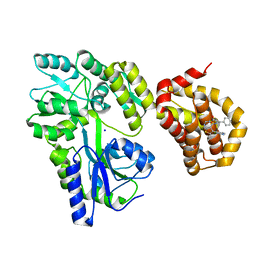 | | Structure of MBP-Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
4KV3
 
 | |
6OB5
 
 | | Computationally-designed, modular sense/response system (S3-2D) | | Descriptor: | Ankyrin Repeat Domain (AR), S3-2D variant, FARNESYL DIPHOSPHATE, ... | | Authors: | Thompson, M.C, Glasgow, A.A, Huang, Y.M, Fraser, J.S, Kortemme, T. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Computational design of a modular protein sense-response system.
Science, 366, 2019
|
|
4KYD
 
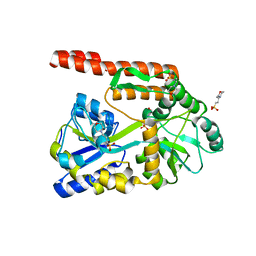 | | Partial Structure of the C-terminal domain of the HPIV4B phosphoprotein, fused to MBP. | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Maltose-binding periplasmic protein, Phosphoprotein, ... | | Authors: | Yegambaram, K, Bulloch, E.M.M, Kingston, R.L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Protein domain definition should allow for conditional disorder.
Protein Sci., 22, 2013
|
|
6D66
 
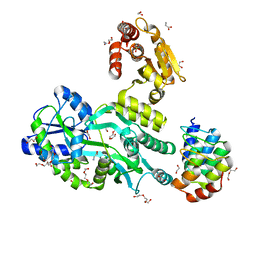 | | Crystal structure of the human dual specificity 1 catalytic domain (C258S) as a maltose binding protein fusion in complex with the designed AR protein mbp3_16 | | Descriptor: | 1,2-ETHANEDIOL, D-ALANINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gumpena, R, Waugh, D.S, Lountos, G.T. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4OGM
 
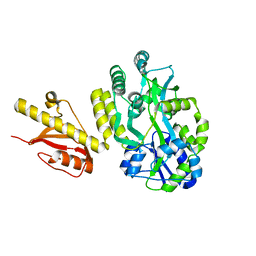 | | MBP-fusion protein of PilA1 residues 26-159 | | Descriptor: | Maltose ABC transporter periplasmic protein, pilin protein chimera, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
1SVX
 
 | | Crystal structure of a designed selected Ankyrin Repeat protein in complex with the Maltose Binding Protein | | Descriptor: | Ankyrin Repeat Protein off7, Maltose-binding periplasmic protein | | Authors: | Binz, H.K, Amstutz, P, Kohl, A, Stumpp, M.T, Briand, C, Forrer, P, Gruetter, M.G, Plueckthun, A. | | Deposit date: | 2004-03-30 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | High-affinity binders selected from designed ankyrin repeat protein libraries
NAT.BIOTECHNOL., 22, 2004
|
|
5GXT
 
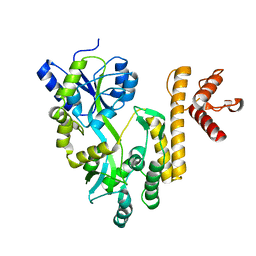 | | Crystal structure of PigG | | Descriptor: | MAGNESIUM ION, Maltose-binding periplasmic protein,PigG | | Authors: | Zhang, F, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2016-09-20 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Crystal structure of MBP-PigG fusion protein and the essential function of PigG in the prodigiosin biosynthetic pathway in Serratia marcescens FS14.
Int. J. Biol. Macromol., 99, 2017
|
|
4XR8
 
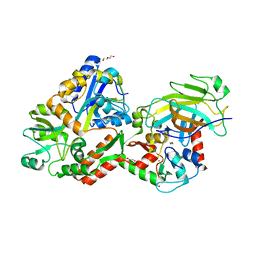 | | Crystal structure of the HPV16 E6/E6AP/p53 ternary complex at 2.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Martinez-Zapien, D, Ruiz, F.X, Mitschler, A, Podjarny, A, Trave, G, Zanier, K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Nature, 529, 2016
|
|
4WTH
 
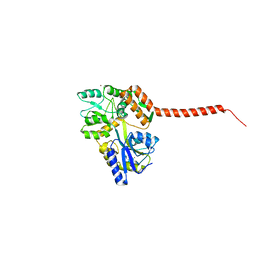 | | Ataxin-3 Carboxy Terminal Region - Crystal C2 (triclinic) | | Descriptor: | Maltose-binding periplasmic protein, Ataxin-3 chimera, ZINC ION, ... | | Authors: | Zhemkov, V.A, Kim, M. | | Deposit date: | 2014-10-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The 2.2-Angstrom resolution crystal structure of the carboxy-terminal region of ataxin-3.
FEBS Open Bio, 6, 2016
|
|
3HST
 
 | |
7DDE
 
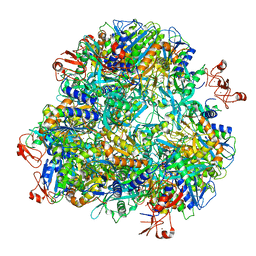 | | Cryo-EM structure of the Ape4 and Nbr1 complex | | Descriptor: | Aspartyl aminopeptidase 1,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
4JKM
 
 | |
6ANV
 
 | | Crystal structure of anti-CRISPR protein AcrF1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
