5KA5
 
 | |
3MNZ
 
 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment with a gp41 MPER-derived peptide bearing Ala substitutions in a helical conformation | | Descriptor: | ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, SODIUM ION, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
1GZL
 
 | | Crystal structure of C14linkmid/IQN17: a cross-linked inhibitor of HIV-1 entry bound to the gp41 hydrophobic pocket | | Descriptor: | CHLORIDE ION, ENVELOPE GLYCOPROTEIN GP41, FUSION PROTEIN BETWEEN THE HYDROPHOBIC POCKET OF HIV GP41 AND GENERAL CONTROL PROTEIN GCN4-PIQI, ... | | Authors: | Sia, S.K, Carr, P.A, Cochran, A.G, Malashkevich, V.M, Kim, P.S. | | Deposit date: | 2002-05-23 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Short Constrained Peptides that Inhibit HIV-1 Entry
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5CIL
 
 | | Crystal Structure of non-neutralizing version of 4E10 (WDWD) with epitope bound | | Descriptor: | ACETATE ION, CHLORIDE ION, FAB 4E10 HEAVY CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
5KA6
 
 | |
1K34
 
 | | Crystal structure analysis of gp41 core mutant | | Descriptor: | Transmembrane glycoprotein GP41 | | Authors: | Shu, W, Lu, M. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion.
Biochemistry, 41, 2002
|
|
6PDS
 
 | | Vaccine-elicited NHP FP-targeting antibody 0PV-a.04 in complex with HIV fusion peptide (residue 512-519) | | Descriptor: | HIV-1 fusion peptide residue 512-519, SULFATE ION, antibody 0PV-a.04 heavy chain, ... | | Authors: | Xu, K, Liu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Modular recognition of antigens provides a mechanism that improves vaccine-elicited antibody-class frequencies
To Be Published
|
|
5Z0W
 
 | |
3VTP
 
 | | HIV fusion inhibitor MT-C34 | | Descriptor: | Transmembrane protein gp41 | | Authors: | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | Deposit date: | 2012-06-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The M-T hook structure is critical for design of HIV-1 fusion inhibitors.
J.Biol.Chem., 287, 2012
|
|
6R2G
 
 | | Crystal structure of a single-chain protein mimetic of the gp41 NHR trimer in complex with the synthetic CHR peptide C34 | | Descriptor: | Envelope glycoprotein gp160, PHOSPHATE ION, Single-chain protein mimetics of the N-terminal heptad-repeat region of gp41 | | Authors: | Camara-Artigas, A, Conejero-Lara, F, Jurado, S, Cano-Munoz, M, Morel, B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Thermodynamic Analysis of HIV-1 Fusion Inhibition Using Small gp41 Mimetic Proteins.
J.Mol.Biol., 431, 2019
|
|
5CN0
 
 | | Artificial HIV fusion inhibitor AP2 fused to the C-terminus of gp41 NHR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein,AP2, MAGNESIUM ION | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
6UBI
 
 | |
6PDU
 
 | | Vaccine-elicited NHP FP-targeting antibody 13N024-a.01 in complex with HIV fusion peptide (residue 512-519) | | Descriptor: | HIV-1 fusion peptide residue 512-519, antibody 13N024-a.01, heavy chain, ... | | Authors: | Xu, K, Liu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Modular recognition of antigens provides a mechanism that improves vaccine-elicited antibody-class frequencies
To Be Published
|
|
6O3L
 
 | |
6MQC
 
 | |
8F3B
 
 | |
7N08
 
 | |
5YC0
 
 | | Crystal structure of LP-46/N44 | | Descriptor: | Envelope glycoprotein, LP-46 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus
J. Biol. Chem., 293, 2018
|
|
2CMR
 
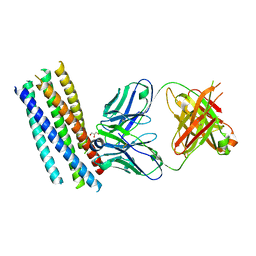 | | Crystal structure of the HIV-1 neutralizing antibody D5 Fab bound to the gp41 inner-core mimetic 5-helix | | Descriptor: | D5, GLYCEROL, TRANSMEMBRANE GLYCOPROTEIN | | Authors: | Luftig, M.A, Mattu, M, Di Giovine, P, Geleziunas, R, Hrin, R, Barbato, G, Bianchi, E, Miller, M.D, Pessi, A, Carfi, A. | | Deposit date: | 2006-05-11 | | Release date: | 2006-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for HIV-1 Neutralization by a Gp41 Fusion Intermediate-Directed Antibody
Nat.Struct.Mol.Biol., 13, 2006
|
|
1DLB
 
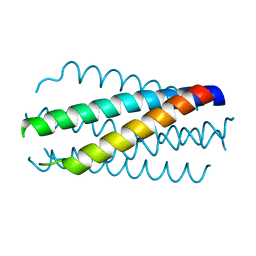 | | HELICAL INTERACTIONS IN THE HIV-1 GP41 CORE REVEALS STRUCTURAL BASIS FOR THE INHIBITORY ACTIVITY OF GP41 PEPTIDES | | Descriptor: | HIV-1 ENVELOPE GLYCOPROTEIN GP41 | | Authors: | Shu, W, Liu, J, Ji, H, Rading, L, Jiang, S, Lu, M. | | Deposit date: | 1999-12-09 | | Release date: | 1999-12-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides.
Biochemistry, 39, 2000
|
|
1AIK
 
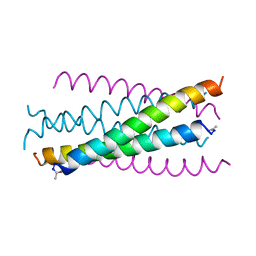 | | HIV GP41 CORE STRUCTURE | | Descriptor: | HIV-1 GP41 GLYCOPROTEIN | | Authors: | Chan, D.C, Fass, D, Berger, J.M, Kim, P.S. | | Deposit date: | 1997-04-20 | | Release date: | 1997-06-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Core structure of gp41 from the HIV envelope glycoprotein.
Cell(Cambridge,Mass.), 89, 1997
|
|
3G9R
 
 | |
2X7R
 
 | |
6PEF
 
 | |
5YB3
 
 | | Crystal structure of HP23L/N36 | | Descriptor: | Envelope glycoprotein, HP23L | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
