1MIL
 
 | | TRANSFORMING PROTEIN | | Descriptor: | SHC ADAPTOR PROTEIN | | Authors: | Mikol, V. | | Deposit date: | 1995-09-20 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the SH2 domain from the adaptor protein SHC: a model for peptide binding based on X-ray and NMR data.
J.Mol.Biol., 254, 1995
|
|
1MIM
 
 | | IGG FAB FRAGMENT (CD25-BINDING) | | Descriptor: | CHIMERIC SDZ CHI621 | | Authors: | Mikol, V. | | Deposit date: | 1995-12-04 | | Release date: | 1997-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the fab fragment of SDZ CHI621: a chimeric antibody against CD25.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1MIO
 
 | | X-RAY CRYSTAL STRUCTURE OF THE NITROGENASE MOLYBDENUM-IRON PROTEIN FROM CLOSTRIDIUM PASTEURIANUM AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | Authors: | Kim, J, Woo, D, Rees, D.C. | | Deposit date: | 1993-03-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of the nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 3.0-A resolution.
Biochemistry, 32, 1993
|
|
1MIQ
 
 | | Crystal structure of proplasmepsin from the human malarial pathogen Plasmodium vivax | | Descriptor: | plasmepsin | | Authors: | Bernstein, N.K, Cherney, M.M, Yowell, C.A, Dame, J.B, James, M.N. | | Deposit date: | 2002-08-23 | | Release date: | 2002-09-18 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation of P. vivax plasmepsin.
J.Mol.Biol., 329, 2003
|
|
1MIR
 
 | | RAT PROCATHEPSIN B | | Descriptor: | PROCATHEPSIN B | | Authors: | Cygler, M, Sivaraman, J, Grochulski, P, Coulombe, R, Storer, A.C, Mort, J.S. | | Deposit date: | 1996-01-12 | | Release date: | 1997-01-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion.
Structure, 4, 1996
|
|
1MIS
 
 | |
1MIT
 
 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
1MIU
 
 | | Structure of a BRCA2-DSS1 complex | | Descriptor: | Breast Cancer type 2 susceptibility protein, Deleted in split hand/split foot protein 1, MERCURY (II) ION | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-23 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA
structure
Science, 297, 2002
|
|
1MIV
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme | | Descriptor: | MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIX
 
 | | Crystal structure of a FERM domain of Talin | | Descriptor: | Talin | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
1MIY
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIZ
 
 | | Crystal structure of an integrin beta3-talin chimera | | Descriptor: | TALIN, integrin beta3 | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
1MJ0
 
 | | SANK E3_5: an artificial Ankyrin repeat protein | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SANK E3_5 Protein, SULFATE ION | | Authors: | Kohl, A, Binz, H.K, Forrer, P, Stumpp, M.T, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2002-08-26 | | Release date: | 2003-01-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Designed to be stable: Crystal structure of a consensus ankyrin repeat protein
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1MJ1
 
 | | FITTING THE TERNARY COMPLEX OF EF-Tu/tRNA/GTP AND RIBOSOMAL PROTEINS INTO A 13 A CRYO-EM MAP OF THE COLI 70S RIBOSOME | | Descriptor: | Elongation Factor Tu, L11 ribosomal protein, Phe-tRNA, ... | | Authors: | Stark, H, Rodnina, M.V, Wieden, H.-J, Zemlin, F, Wintermeyer, W, Vanheel, M. | | Deposit date: | 2002-08-26 | | Release date: | 2002-11-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Ribosome Interactions of Aminoacyl-tRNA and Elongation Factor TU in the
Codon Recognition Complex
Nat.Struct.Biol., 9, 2002
|
|
1MJ2
 
 | |
1MJ3
 
 | | Crystal Structure Analysis of rat enoyl-CoA hydratase in complex with hexadienoyl-CoA | | Descriptor: | ENOYL-COA HYDRATASE, MITOCHONDRIAL, HEXANOYL-COENZYME A | | Authors: | Bell, A.F, Feng, Y, Hofstein, H.A, Parikh, S, Wu, J, Rudolph, M.J, Kisker, C, Tonge, P.J. | | Deposit date: | 2002-08-26 | | Release date: | 2002-09-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stereoselectivity of Enoyl-CoA Hydratase Results from Preferential Activation of
One of Two Bound Substrate Conformers
Chem.Biol., 9, 2002
|
|
1MJ4
 
 | | Crystal Structure Analysis of the cytochrome b5 domain of human sulfite oxidase | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Rudolph, M.J, Johnson, J.L, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2002-08-26 | | Release date: | 2002-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A structure of the human sulfite oxidase cytochrome b(5) domain.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MJ5
 
 | | LINB (haloalkane dehalogenase) from sphingomonas paucimobilis UT26 at atomic resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Oakley, A.J, Damborsky, J, Wilce, M.C. | | Deposit date: | 2002-08-27 | | Release date: | 2003-08-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26 at 0.95 A resolution: dynamics of catalytic residues.
Biochemistry, 43, 2004
|
|
1MJ7
 
 | | Crystal Structure Of The Complex Of The Fab fragment of Esterolytic Antibody MS5-393 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS5-393, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-27 | | Release date: | 2003-09-23 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MJ8
 
 | | High Resolution Crystal Structure Of The Fab Fragment of The Esterolytic Antibody MS6-126 | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN MS6-126, PHOSPHATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-27 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MJ9
 
 | | Crystal structure of yeast Esa1(C304S) mutant complexed with Coenzyme A | | Descriptor: | COENZYME A, ESA1 PROTEIN, SODIUM ION | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
1MJA
 
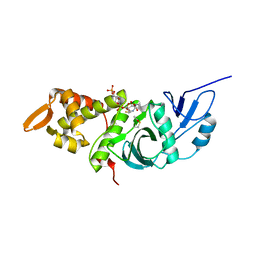 | | Crystal structure of yeast Esa1 histone acetyltransferase domain complexed with acetyl coenzyme A | | Descriptor: | COENZYME A, Esa1 protein | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
1MJB
 
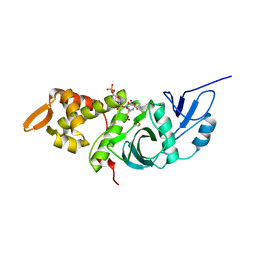 | | Crystal structure of yeast Esa1 histone acetyltransferase E338Q mutant complexed with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, Esa1 protein | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
1MJC
 
 | |
