3R5T
 
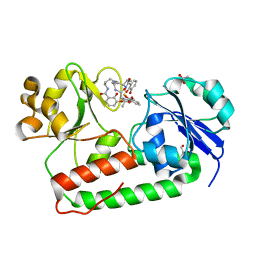 | | Crystal structure of holo-ViuP | | Descriptor: | (4S,5R)-N-{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-N-[3-({[(4S,5R)-2-(2,3-dihydroxyphenyl)-5-met hyl-4,5-dihydro-1,3-oxazol-4-yl]carbonyl}amino)propyl]-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
4QKX
 
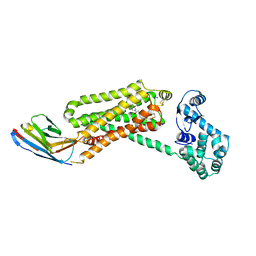 | | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody | | Descriptor: | 4-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(2-sulfanylethoxy)phenyl]ethyl}amino)ethyl]benzene-1,2-diol, Beta-2 adrenergic receptor, R9 protein, ... | | Authors: | Weichert, D, Kruse, A.C, Manglik, A, Hiller, C, Zhang, C, Huebner, H, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Covalent agonists for studying G protein-coupled receptor activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8JNC
 
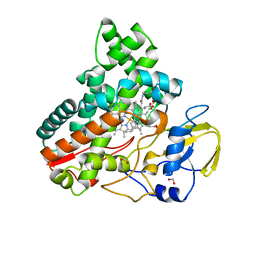 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-maltophilin | | Descriptor: | (1Z,3E,5S,8R,9S,10S,11R,13R,15R,16S,18Z,24S,25S)-11-ethyl-2,24-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-7,20,27,28-tetraone, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JNQ
 
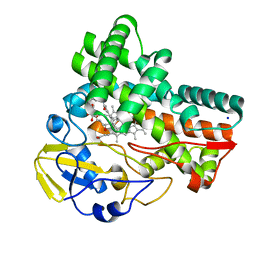 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with a substrate compound c | | Descriptor: | (1Z,3E,5E,7S,8R,10S,11R,13R,15R,16E,18E,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazatetracyclo[23.2.1.09,13.08,15]octacosa-1(2),3,5,16,18-pentaene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JOO
 
 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate ikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-08 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JNO
 
 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-deOH-HSAF | | Descriptor: | (1Z,3E,5S,7S,8R,9S,10S,11R,13R,15R,16S,18Z,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4GO3
 
 | |
3Q7J
 
 | | Engineered Thermoplasma Acidophilum F3 factor mimics human aminopeptidase N (APN) as a target for anticancer drug development | | Descriptor: | L-phenylalanyl-N6-[(benzyloxy)carbonyl]-N1-hydroxy-L-lysinamide, Tricorn protease-interacting factor F3, ZINC ION | | Authors: | Su, J, Wang, Q, Feng, J, Zhang, C, Zhu, D, We, T, Xu, W, Gu, L. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Engineered Thermoplasma acidophilum factor F3 mimics human aminopeptidase N (APN) as a target for anticancer drug development
Bioorg.Med.Chem., 19, 2011
|
|
4DOY
 
 | | Crystal structure of Dibenzothiophene desulfurization enzyme C | | Descriptor: | Dibenzothiophene desulfurization enzyme C, GLYCEROL | | Authors: | Liu, S, Zhang, C, Zhu, D, Gu, L. | | Deposit date: | 2012-02-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of DszC from Rhodococcus sp. XP at 1.79 angstrom
Proteins, 82, 2014
|
|
8JNP
 
 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with the substrate ikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450 CftA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5Z3Q
 
 | | Crystal Structure of a Soluble Fragment of Poliovirus 2C ATPase (2.55 Angstrom) | | Descriptor: | PHOSPHATE ION, PV-2C, ZINC ION | | Authors: | Guan, H, Tian, J, Zhang, C, Qin, B, Cui, S. | | Deposit date: | 2018-01-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Crystal structure of a soluble fragment of poliovirus 2CATPase
PLoS Pathog., 14, 2018
|
|
7V1U
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZJ12 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(3-ethyl-6-methoxy-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide | | Authors: | Zhang, M, Wang, C, Zhang, C, Zhang, Y, Xu, Y. | | Deposit date: | 2021-08-06 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design, synthesis and pharmacological characterization of N-(3-ethylbenzo[d]isoxazol-5-yl) sulfonamide derivatives as BRD4 inhibitors against acute myeloid leukemia.
Acta Pharmacol.Sin., 43, 2022
|
|
7V2J
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 33 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(3-ethyl-6-methoxy-1,2-benzoxazol-5-yl)-4-methoxy-benzenesulfonamide | | Authors: | Zhang, M, Wang, C, Zhang, C, Zhang, Y, Xu, Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Design, synthesis and pharmacological characterization of N-(3-ethylbenzo[d]isoxazol-5-yl) sulfonamide derivatives as BRD4 inhibitors against acute myeloid leukemia.
Acta Pharmacol.Sin., 43, 2022
|
|
6KO0
 
 | | The crystal structue of PDE10A complexed with 1i | | Descriptor: | 3-[2-(5-methyl-1-phenyl-benzimidazol-2-yl)ethyl]chromen-4-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, Yu, Y.F, Zhang, C, Guo, L, Wu, D, Luo, H.-B. | | Deposit date: | 2019-08-07 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.600029 Å) | | Cite: | Discovery and Optimization of Chromone Derivatives as Novel Selective Phosphodiesterase 10 Inhibitors.
Acs Chem Neurosci, 11, 2020
|
|
6KO1
 
 | | The crystal structue of PDE10A complexed with 2d | | Descriptor: | 6-chloranyl-3-[2-(5-methyl-1-phenyl-benzimidazol-2-yl)ethyl]chromen-4-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, Yu, Y.F, Zhang, C, Guo, L, Wu, D, Luo, H.-B. | | Deposit date: | 2019-08-07 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Optimization of Chromone Derivatives as Novel Selective Phosphodiesterase 10 Inhibitors.
Acs Chem Neurosci, 11, 2020
|
|
7V9M
 
 | | Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Chen, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6AIJ
 
 | | Cyclodextrin glycosyltransferase from Paenibacillus macerans mutant N603D | | Descriptor: | CALCIUM ION, Cyclomaltodextrin glucanotransferase | | Authors: | Li, C.M, Ban, X.F, Li, Z.F, Li, Y.L, Cheng, S.D, Zhang, C.Y, Jin, T.C, Gu, Z.B. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Cyclodextrin glycosyltransferase from Paenibacillus macerans mutant N603D
To Be Published
|
|
6ODC
 
 | | Crystal structure of HDAC8 in complex with compound 30 | | Descriptor: | (2E)-3-[2-(3-cyclopentyl-5,5-dimethyl-2-oxoimidazolidin-1-yl)phenyl]-N-hydroxyprop-2-enamide, 1,2-ETHANEDIOL, Histone deacetylase 8, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6ODA
 
 | | Crystal structure of HDAC8 in complex with compound 2 | | Descriptor: | Histone deacetylase 8, N-{2-[3-(hydroxyamino)-3-oxopropyl]phenyl}-3-(trifluoromethyl)benzamide, POTASSIUM ION, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
7V9L
 
 | | Cryo-EM structure of the SV1-Gs complex. | | Descriptor: | GHRH receptor splice variant 1,GHRH receptor splice variant 1,GHRH receptor splice variant 1,SV1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Cheng, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6NSQ
 
 | | Crystal structure of BRAF kinase domain bound to the inhibitor 2l | | Descriptor: | 5-[(4-amino-1-ethyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)ethynyl]-N-(4-chlorophenyl)-6-methylisoquinolin-1-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Zhang, C, Sicheri, F. | | Deposit date: | 2019-01-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rigidification Dramatically Improves Inhibitor Selectivity for RAF Kinases.
Acs Med.Chem.Lett., 10, 2019
|
|
4GO4
 
 | | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative gamma-hydroxymuconic semialdehyde dehydrogenase | | Authors: | Su, J, Zhang, C, Liu, S, Zhu, D, Gu, L. | | Deposit date: | 2012-08-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide
To be Published
|
|
9BJK
 
 | | Inactive mu opioid receptor bound to Nb6, naloxone and NAM | | Descriptor: | Mu-type opioid receptor, Naloxone, Nalpha-[({(1M)-1-[5-(benzyloxy)pyridin-3-yl]naphthalen-2-yl}sulfanyl)acetyl]-3-methoxy-N,4-dimethyl-L-phenylalaninamide, ... | | Authors: | O'Brien, E.S, Wang, H, Kaavya Krishna, K, Zhang, C, Kobilka, B.K. | | Deposit date: | 2024-04-25 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | A mu-opioid receptor modulator that works cooperatively with naloxone.
Nature, 631, 2024
|
|
3R5S
 
 | | Crystal structure of apo-ViuP | | Descriptor: | Ferric vibriobactin ABC transporter, periplasmic ferric vibriobactin-binding protein | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2012-05-30 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
