3OMA
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OMN
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5GP7
 
 | | Structural basis for the binding between Tankyrase-1 and USP25 | | Descriptor: | GLYCEROL, Tankyrase-1, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Liu, J, Xu, D, Fu, T, Pan, L. | | Deposit date: | 2016-08-01 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | USP25 regulates Wnt signaling by controlling the stability of tankyrases
Genes Dev., 31, 2017
|
|
3OM3
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OXS
 
 | | Crystal Structure of HLA A*02:07 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-22 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
3OXR
 
 | | Crystal Structure of HLA A*02:06 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
4HBK
 
 | | Structure of the Aldose Reductase from Schistosoma japonicum | | Descriptor: | Aldo-keto reductase family 1, member B4 (Aldose reductase) | | Authors: | Liu, J, Cheng, J, Zhang, X, Yang, Z, Hu, W, Xu, Y. | | Deposit date: | 2012-09-28 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aldose reductase from Schistosoma japonicum: crystallization and structure-based inhibitor screening for discovering antischistosomal lead compounds.
Parasit Vectors, 6, 2013
|
|
3N27
 
 | |
1Z0Z
 
 | | Crystal structure of a NAD kinase from Archaeoglobus fulgidus in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable inorganic polyphosphate/ATP-NAD kinase | | Authors: | Liu, J, Lou, Y, Yokota, H, Adams, P.D, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-03-02 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of an NAD Kinase from Archaeoglobus fulgidus in Complex with ATP, NAD, or NADP
J.Mol.Biol., 354, 2005
|
|
1Z0U
 
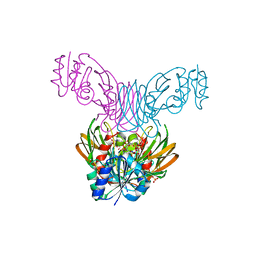 | | Crystal structure of a NAD kinase from Archaeoglobus fulgidus bound by NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable inorganic polyphosphate/ATP-NAD kinase, SULFATE ION | | Authors: | Liu, J, Lou, Y, Yokota, H, Adams, P.D, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-03-02 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of an NAD Kinase from Archaeoglobus fulgidus in Complex with ATP, NAD, or NADP
J.Mol.Biol., 354, 2005
|
|
3OX8
 
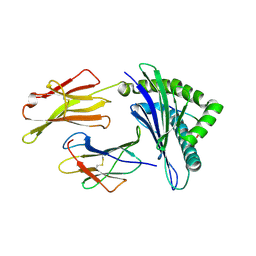 | | Crystal Structure of HLA A*02:03 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
3GWO
 
 | | Structure of the C-terminal Domain of a Putative HIV-1 gp41 Fusion Intermediate | | Descriptor: | Envelope glycoprotein gp160, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), SODIUM ION | | Authors: | Liu, J. | | Deposit date: | 2009-04-01 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Role of a putative gp41 dimerization domain in human immunodeficiency virus type 1 membrane fusion.
J.Virol., 84, 2010
|
|
3H01
 
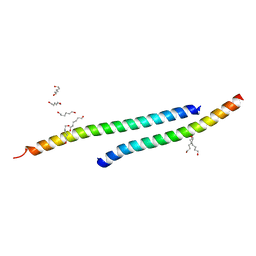 | |
3H00
 
 | |
3TO2
 
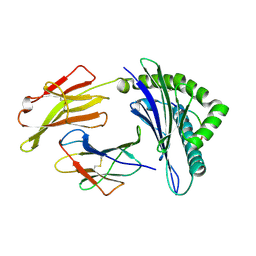 | | Structure of HLA-A*0201 complexed with peptide Md3-C9 derived from a clustering region of restricted cytotoxic T lymphocyte epitope from SARS-CoV M protein | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Md3-C9 peptide derived from Membrane glycoprotein | | Authors: | Liu, J, Qi, J, Gao, F, Yan, J, Gao, G.F. | | Deposit date: | 2011-09-03 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional and Structural Definition of a Clustering Region of HLA-A2-restricted Cytotoxic T Lymphocyte Epitopes
Sci.Technology Rev., 29, 2011
|
|
4IDV
 
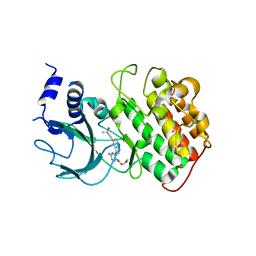 | | Crystal Structure of NIK with compound 4-{3-[2-amino-5-(2-methoxyethoxy)pyrimidin-4-yl]-1H-indol-5-yl}-2-methylbut-3-yn-2-ol (13V) | | Descriptor: | 4-{3-[2-amino-5-(2-methoxyethoxy)pyrimidin-4-yl]-1H-indol-5-yl}-2-methylbut-3-yn-2-ol, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Liu, J, Sudom, A, Wang, Z. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibiting NF-KB-inducing kinase (NIK): Discovery, structure-based design, synthesis, structure activity relationship, and co-crystal structures
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4CQZ
 
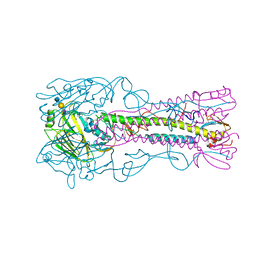 | | Crystal Structure of H5 (VN1194) Gln196Arg Mutant Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Collins, P.J, Vachieri, S.G, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
4QKT
 
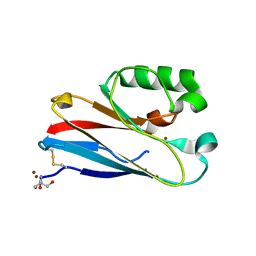 | | Azurin mutant M121EM44K with copper | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Azurin, ... | | Authors: | Liu, J, Robinson, H, Lu, Y. | | Deposit date: | 2014-06-09 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Redesigning the Blue Copper Azurin into a Redox-Active Mononuclear Nonheme Iron Protein: Preparation and Study of Fe(II)-M121E Azurin.
J.Am.Chem.Soc., 136, 2014
|
|
4IDT
 
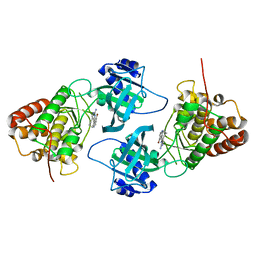 | | Crystal Structure of NIK with 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine (T28) | | Descriptor: | 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Liu, J, Sudom, A, Wang, Z. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibiting NF-KB-inducing kinase (NIK): Discovery, structure-based design, synthesis, structure activity relationship, and co-crystal structures
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3TCP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC569 | | Descriptor: | 1-[(trans-4-aminocyclohexyl)methyl]-N-butyl-3-(4-fluorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-6-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, J, Yang, C, Simpson, C, DeRyckere, D, Van Deusen, A, Miley, M, Kireev, D.B, Norris-Drouin, J, Sather, S, Hunter, D, Patel, H.S, Janzen, W.P, Machius, M, Johnson, G, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia.
ACS Med Chem Lett, 3, 2012
|
|
4QLW
 
 | | Azurin mutant M121E with iron | | Descriptor: | Azurin, FE (III) ION, NITRATE ION, ... | | Authors: | Liu, J, Robinson, H, Lu, Y. | | Deposit date: | 2014-06-13 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redesigning the Blue Copper Azurin into a Redox-Active Mononuclear Nonheme Iron Protein: Preparation and Study of Fe(II)-M121E Azurin.
J.Am.Chem.Soc., 136, 2014
|
|
4F0I
 
 | | Crystal structure of apo TrkA | | Descriptor: | High affinity nerve growth factor receptor | | Authors: | Liu, J. | | Deposit date: | 2012-05-04 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Crystal Structures of TrkA and TrkB Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
4FQG
 
 | | Crystal structure of the TCERG1 FF4-6 tandem repeat domain | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Transcription elongation regulator 1 | | Authors: | Liu, J, Fan, S, Lee, C.J, Greenleaf, A.L, Zhou, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Interaction of the Transcription Elongation Regulator TCERG1 with RNA Polymerase II Requires Simultaneous Phosphorylation at Ser2, Ser5, and Ser7 within the Carboxyl-terminal Domain Repeat.
J.Biol.Chem., 288, 2013
|
|
5WWD
 
 | | Crystal structure of AtNUDX1 | | Descriptor: | AMMONIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2016-12-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5WY6
 
 | | Crystal structure of AtNUDX1 (E56A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nudix hydrolase 1, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
