1R28
 
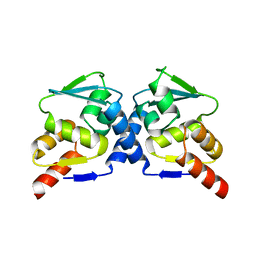 | | Crystal Structure of the B-Cell Lymphoma 6 (BCL6) BTB domain to 2.2 Angstrom | | Descriptor: | B-cell lymphoma 6 protein | | Authors: | Ahmad, K.F, Melnick, A, Lax, S.A, Bouchard, D, Liu, J, Kiang, C.L, Mayer, S, Licht, J.D, Prive, G.G. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain.
Mol.Cell, 12, 2003
|
|
2VF2
 
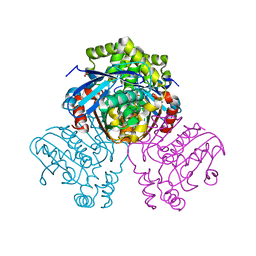 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1RF2
 
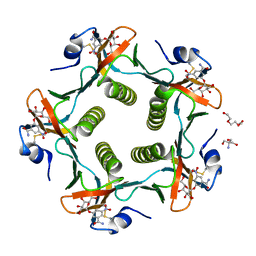 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV4 | | Descriptor: | 1,3-BIS-([3-[3-[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBUTENYL]-AMINO-PROPOXY-ETHOXY-ETHOXY]-PROPYL-]AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-07 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1Z26
 
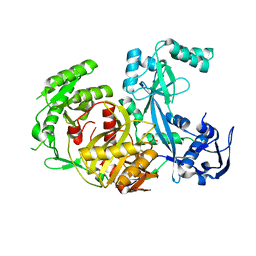 | | Structure of Pyrococcus furiosus Argonaute with bound tungstate | | Descriptor: | Argonaute, TUNGSTATE(VI)ION | | Authors: | Rivas, F.V, Tolia, N.H, Song, J.J, Aragon, J.P, Liu, J, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Purified Argonaute2 and an siRNA form recombinant human RISC.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1Z25
 
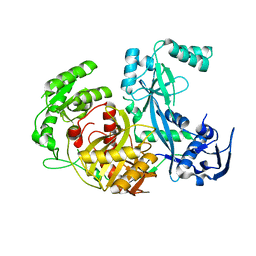 | | Structure of P.furiosus Argonaute with bound Mn2+ | | Descriptor: | Argonaute, MANGANESE (II) ION | | Authors: | Rivas, F.V, Tolia, N.H, Song, J.J, Aragon, J.P, Liu, J, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Purified Argonaute2 and an siRNA form recombinant human RISC.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1T8U
 
 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP and tetrasaccharide substrate | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
4UC1
 
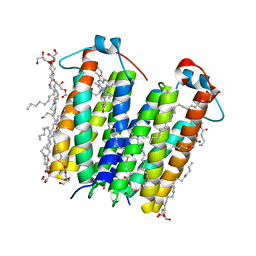 | | High resolution crystal structure of translocator protein 18kDa (TSPO) from Rhodobacter sphaeroides (A139T Mutant) in C2 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, METHOXY-ETHOXYL, ... | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
5ZMD
 
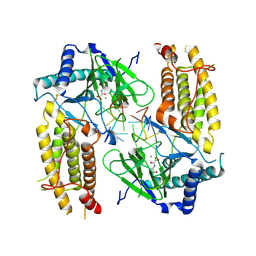 | | Crystal structure of FTO in complex with m6dA modified ssDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, DNA (5'-D(P*TP*CP*TP*(6MA)P*TP*AP*TP*CP*G)-3'), MANGANESE (II) ION, ... | | Authors: | Zhang, X, Wei, L.H, Luo, J, Xiao, Y, Liu, J, Zhang, W, Zhang, L, Jia, G.F. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4UC3
 
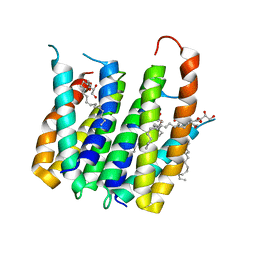 | | Translocator protein 18 kDa (TSPO) from Rhodobacter sphaeroides wild type | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, TRANSLOCATOR PROTEIN TSPO | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
1TU4
 
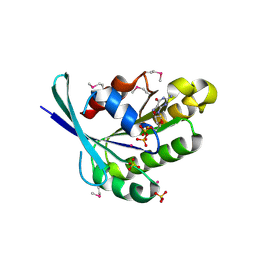 | | Crystal Structure of Rab5-GDP Complex | | Descriptor: | COBALT (II) ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-5A, ... | | Authors: | Zhu, G, Zhai, P, Liu, J, Terzyan, S, Li, G, Zhang, X.C. | | Deposit date: | 2004-06-24 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of Rab5-Rabaptin5 interaction in endocytosis
Nat.Struct.Mol.Biol., 11, 2004
|
|
4UC2
 
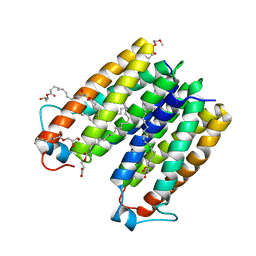 | | Crystal structure of translocator protein 18kDa (TSPO) from rhodobacter sphaeroides (A139T mutant) in P212121 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, TETRAETHYLENE GLYCOL, TRANSLOCATOR PROTEIN TSPO | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
1ZG2
 
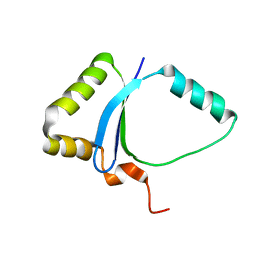 | | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2. | | Descriptor: | Hypothetical UPF0213 protein BH0048 | | Authors: | Aramini, J.M, Swapna, G.V.T, Xiao, R, Ma, L, Shastry, R, Ciano, M, Acton, T.B, Liu, J, Rost, B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-20 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2.
To be Published
|
|
1SQR
 
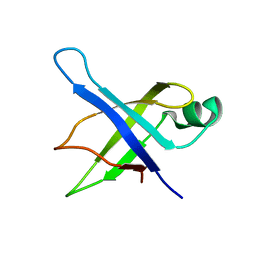 | | Solution Structure of the 50S Ribosomal Protein L35AE from Pyrococcus furiosus. Northeast Structural Genomics Consortium Target PfR48. | | Descriptor: | 50S ribosomal protein L35Ae | | Authors: | Snyder, D.A, Aramini, J.M, Huang, Y.J, Xiao, R, Cort, J.R, Shastry, R, Ma, L.C, Liu, J, Rost, B, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 50S Ribosomal Protein L35AE from Pyrococcus furiosus: Northeast Structural Genomics Consortium Target PfR48
To be Published
|
|
1T8T
 
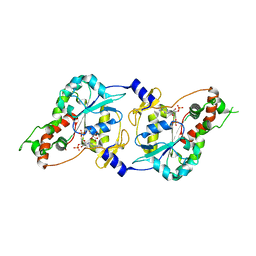 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CITRIC ACID, heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1 | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
2A5I
 
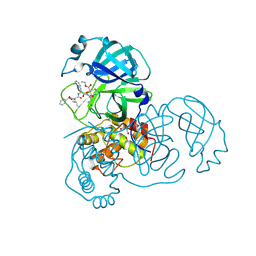 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in the space group C2 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 1,2-ETHANEDIOL, 3C-like peptidase, ... | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5K
 
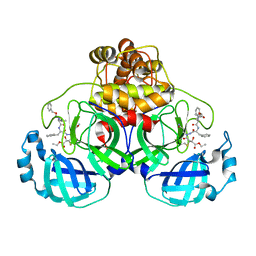 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in space group P212121 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like peptidase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5A
 
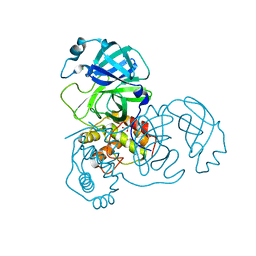 | | Crystal structure of unbound SARS coronavirus main peptidase in the space group C2 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like peptidase, CHLORIDE ION | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
3DJB
 
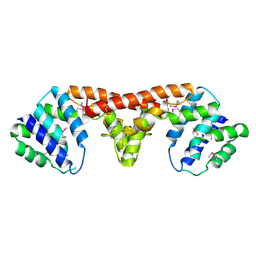 | | Crystal structure of a HD-superfamily hydrolase (BT9727_1981) from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR114 | | Descriptor: | Hydrolase, HD family, MAGNESIUM ION | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Vorobiev, S.M, Janjua, H, Fang, Y, Xiao, R, Cunningham, K, Maglaqui, M, Owen, L.A, Wang, D, Baran, M.C, Liu, J, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-23 | | Release date: | 2008-08-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a HD-superfamily hydrolase (BT9727_1981) from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR114
To be Published
|
|
8GJW
 
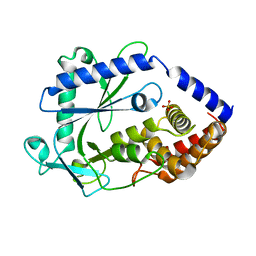 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
3ERJ
 
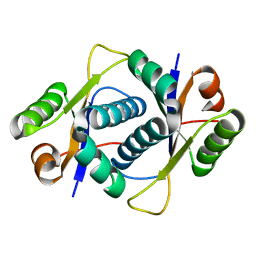 | | Crystal structure of the peptidyl-tRNA hydrolase AF2095 from Archaeglobus fulgidis. Northeast Structural Genomics Consortium target GR4 | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Conover, K, Janjua, H, Xiao, R, Cunningham, K, Ma, L.-C, Cooper, B, Baran, M.C, Liu, J, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-02 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the peptidyl-tRNA hydrolase AF2095 from Archaeglobus fulgidis. Northeast Structural Genomics Consortium target GR4
To be Published
|
|
3E5Z
 
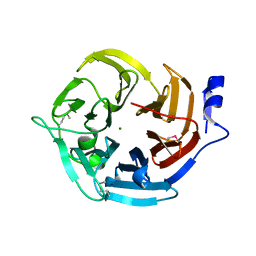 | | X-Ray structure of the putative gluconolactonase in protein family PF08450. Northeast Structural Genomics Consortium target DrR130. | | Descriptor: | MAGNESIUM ION, putative Gluconolactonase | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Tong, S.N, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | X-Ray structure of the putative gluconolactonase in protein family PF08450. Northeast Structural Genomics Consortium target DrR130.
To be Published
|
|
7KBG
 
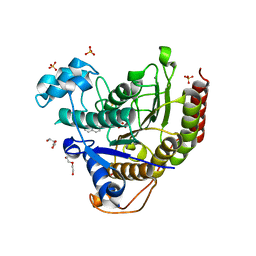 | |
4EJQ
 
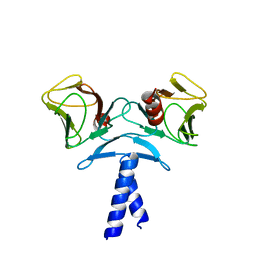 | | Crystal structure of KIF1A C-CC1-FHA | | Descriptor: | Kinesin-like protein KIF1A | | Authors: | Huo, L, Yue, Y, Ren, J, Yu, J, Liu, J, Yu, Y, Ye, F, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-06 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
3FHW
 
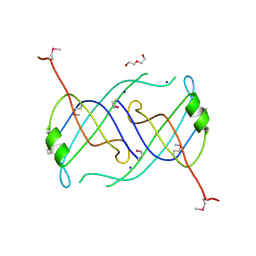 | | Crystal structure of the protein priB from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR162. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Primosomal replication protein n, SODIUM ION | | Authors: | Kuzin, A.P, Neely, H, Seetharaman, J, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-10 | | Release date: | 2008-12-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein priB from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR162.
To be Published
|
|
3CER
 
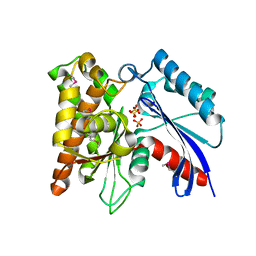 | | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13 | | Descriptor: | Possible exopolyphosphatase-like protein, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Chen, Y, Neely, H, Seetharaman, J, Shastry, R, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-04-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13.
To be Published
|
|
