3TO2
 
 | | Structure of HLA-A*0201 complexed with peptide Md3-C9 derived from a clustering region of restricted cytotoxic T lymphocyte epitope from SARS-CoV M protein | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Md3-C9 peptide derived from Membrane glycoprotein | | Authors: | Liu, J, Qi, J, Gao, F, Yan, J, Gao, G.F. | | Deposit date: | 2011-09-03 | | Release date: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional and Structural Definition of a Clustering Region of HLA-A2-restricted Cytotoxic T Lymphocyte Epitopes
Sci.Technology Rev., 29, 2011
|
|
4NT6
 
 | | HLA-C*0801 Crystal Structure | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, Cw-8 alpha chain, ... | | Authors: | Liu, J.X, Toh, X.Y, Ren, E.C. | | Deposit date: | 2013-12-01 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The immunodominant influenza A virus M158-66 cytotoxic T lymphocyte epitope exhibits degenerate class I major histocompatibility complex restriction in humans.
J.Virol., 88, 2014
|
|
3CSE
 
 | | Candida glabrata Dihydrofolate Reductase complexed with NADPH and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J, Anderson, A.C. | | Deposit date: | 2008-04-09 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided development of efficacious antifungal agents targeting Candida glabrata dihydrofolate reductase.
Chem.Biol., 15, 2008
|
|
3TCP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC569 | | Descriptor: | 1-[(trans-4-aminocyclohexyl)methyl]-N-butyl-3-(4-fluorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-6-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, J, Yang, C, Simpson, C, DeRyckere, D, Van Deusen, A, Miley, M, Kireev, D.B, Norris-Drouin, J, Sather, S, Hunter, D, Patel, H.S, Janzen, W.P, Machius, M, Johnson, G, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia.
ACS Med Chem Lett, 3, 2012
|
|
5WEH
 
 | | Cytochrome c oxidase from Rhodobacter sphaeroides in the reduced state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Aa3-type cytochrome c oxidase subunit IV, CALCIUM ION, ... | | Authors: | Liu, J, Ferguson-Miller, F, Ling, Q, Hiser, C. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Role of conformational change and K-path ligands in controlling cytochrome c oxidase activity.
Biochem. Soc. Trans., 45, 2017
|
|
4U88
 
 | |
6LON
 
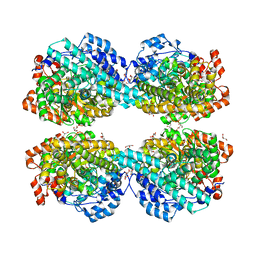 | | Crystal structure of HPSG | | Descriptor: | (2~{S})-2,3-bis(oxidanyl)propane-1-sulfonic acid, GLYCEROL, PFL2/glycerol dehydratase family glycyl radical enzyme, ... | | Authors: | Liu, J, Zhang, Y, Yuchi, Z. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two radical-dependent mechanisms for anaerobic degradation of the globally abundant organosulfur compound dihydroxypropanesulfonate.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
9BA5
 
 | |
9BA4
 
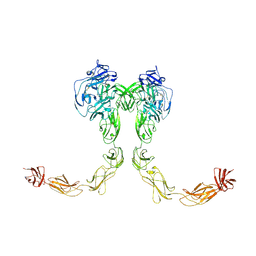 | | Full-length cross-linked Contactin 2 (CNTN2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2 | | Authors: | Liu, J.L, Fan, S.F, Ren, G.R, Rudenko, G.R. | | Deposit date: | 2024-04-03 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of contactin 2 homophilic interaction.
Structure, 2024
|
|
4F0I
 
 | | Crystal structure of apo TrkA | | Descriptor: | High affinity nerve growth factor receptor | | Authors: | Liu, J. | | Deposit date: | 2012-05-04 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Crystal Structures of TrkA and TrkB Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
4FQG
 
 | | Crystal structure of the TCERG1 FF4-6 tandem repeat domain | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Transcription elongation regulator 1 | | Authors: | Liu, J, Fan, S, Lee, C.J, Greenleaf, A.L, Zhou, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Interaction of the Transcription Elongation Regulator TCERG1 with RNA Polymerase II Requires Simultaneous Phosphorylation at Ser2, Ser5, and Ser7 within the Carboxyl-terminal Domain Repeat.
J.Biol.Chem., 288, 2013
|
|
4HBK
 
 | | Structure of the Aldose Reductase from Schistosoma japonicum | | Descriptor: | Aldo-keto reductase family 1, member B4 (Aldose reductase) | | Authors: | Liu, J, Cheng, J, Zhang, X, Yang, Z, Hu, W, Xu, Y. | | Deposit date: | 2012-09-28 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aldose reductase from Schistosoma japonicum: crystallization and structure-based inhibitor screening for discovering antischistosomal lead compounds.
Parasit Vectors, 6, 2013
|
|
4F7P
 
 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009H1N1 PB1 (496-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
2LSJ
 
 | |
2LSG
 
 | |
3EEK
 
 | |
3OX8
 
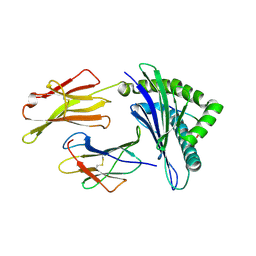 | | Crystal Structure of HLA A*02:03 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
3EEM
 
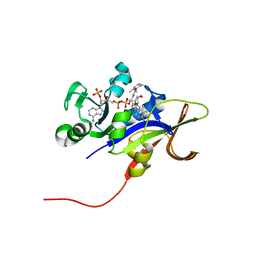 | | Candida glabrata Dihydrofolate Reductase complexed with 2,4-diamino-5-[3-methyl-3-(3-methoxy-5-(2,6-dimethylphenyl)phenyl)prop-1-ynyl]-6-methylpyrimidine(UCP111D26M) and NADPH | | Descriptor: | 5-[(3R)-3-(5-methoxy-2',6'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J, Anderson, A. | | Deposit date: | 2008-09-04 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Probing the active site of Candida glabrata dihydrofolate reductase with high resolution crystal structures and the synthesis of new inhibitors
Chem.Biol.Drug Des., 73, 2009
|
|
3EEL
 
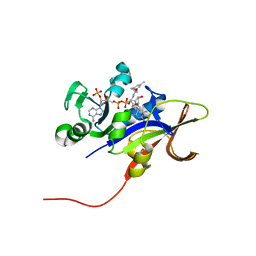 | | Candida glabrata Dihydrofolate Reductase complexed with 2,4-diamino-5-[3-methyl-3-(3-methoxy-5-(3,5-dimethylphenyl)phenyl)prop-1-ynyl]-6-methylpyrimidine(UCP11153TM) and NADPH | | Descriptor: | 5-[(3R)-3-(5-methoxy-3',5'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J, Anderson, A. | | Deposit date: | 2008-09-04 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the active site of Candida glabrata dihydrofolate reductase with high resolution crystal structures and the synthesis of new inhibitors
Chem.Biol.Drug Des., 73, 2009
|
|
3EEJ
 
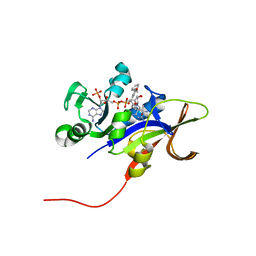 | |
8I5Z
 
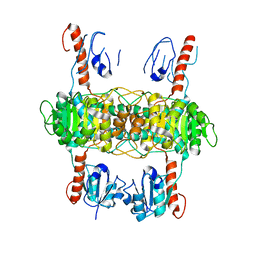 | |
4IDV
 
 | | Crystal Structure of NIK with compound 4-{3-[2-amino-5-(2-methoxyethoxy)pyrimidin-4-yl]-1H-indol-5-yl}-2-methylbut-3-yn-2-ol (13V) | | Descriptor: | 4-{3-[2-amino-5-(2-methoxyethoxy)pyrimidin-4-yl]-1H-indol-5-yl}-2-methylbut-3-yn-2-ol, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Liu, J, Sudom, A, Wang, Z. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibiting NF-KB-inducing kinase (NIK): Discovery, structure-based design, synthesis, structure activity relationship, and co-crystal structures
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3VC4
 
 | |
3VBX
 
 | |
3VBT
 
 | |
