5EE1
 
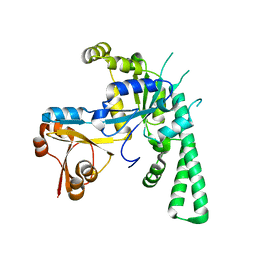 | | Crystal structure of OsYchF1 at pH 7.85 | | Descriptor: | Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EE3
 
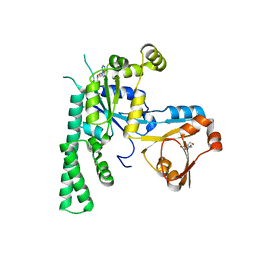 | | COMPLEX STRUCTURE OF OSYCHF1 WITH AMP-PNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5YGI
 
 | |
3NDM
 
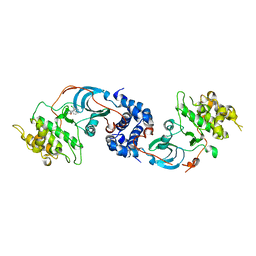 | |
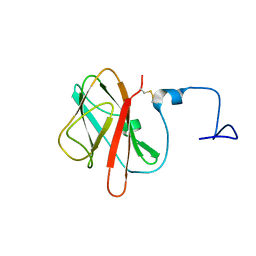
5X5S
 
 | |
2LEF
 
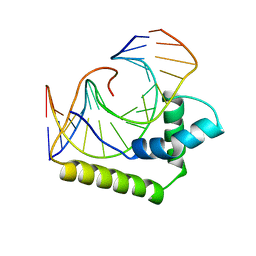 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | Authors: | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
7N4X
 
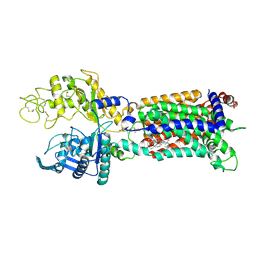 | | Structure of human NPC1L1 mutant-W347R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
7N4U
 
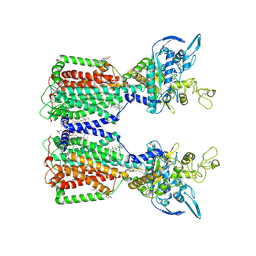 | | Structure of human NPC1L1 | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
7N4V
 
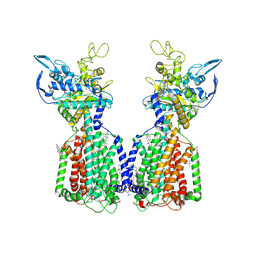 | | Structure of cholesterol-bound human NPC1L1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
7N6R
 
 | | Structure of nevanimibe bound human ACAT2 | | Descriptor: | CHOLESTEROL, OLEIC ACID, Sterol O-acyltransferase 2, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of nevanimibe bound human ACAT2
To Be Published
|
|
7N6Q
 
 | | Structure of PPPA bound human ACAT2 | | Descriptor: | (3S,4R,4aR,6S,6aS,12R,12aS,12bS)-4-[(acetyloxy)methyl]-12-hydroxy-4,6a,12b-trimethyl-11-oxo-9-(pyridin-3-yl)-1,3,4,4a,5,6,6a,12,12a,12b-decahydro-2H,11H-naphtho[2,1-b]pyrano[3,4-e]pyran-3,6-diyl diacetate, CHOLESTEROL, OLEIC ACID, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structure of PPPA bound human ACAT2
To Be Published
|
|
2GNC
 
 | | Crystal structure of srGAP1 SH3 domain in the slit-robo signaling pathway | | Descriptor: | SLIT-ROBO Rho GTPase-activating protein 1 | | Authors: | Li, X, Liu, Y, Gao, F, Bartlam, M, Wu, J.Y, Rao, Z. | | Deposit date: | 2006-04-10 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Robo Proline-rich Motif Recognition by the srGAP1 Src Homology 3 Domain in the Slit-Robo Signaling Pathway
J.Biol.Chem., 281, 2006
|
|
3ID4
 
 | | Crystal Structure of RseP PDZ2 domain fused GKASPV peptide | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ID2
 
 | | Crystal Structure of RseP PDZ2 domain | | Descriptor: | IODIDE ION, Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2FLU
 
 | | Crystal Structure of the Kelch-Neh2 Complex | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 | | Authors: | Li, X, Lo, J, Beamer, L, Hannink, M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling.
Embo J., 25, 2006
|
|
3ID1
 
 | | Crystal Structure of RseP PDZ1 domain | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ID3
 
 | | Crystal Structure of RseP PDZ2 I304A domain | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4HYD
 
 | | Structure of a presenilin family intramembrane aspartate protease in C2221 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
2DC2
 
 | | Solution Structure of PDZ Domain | | Descriptor: | golgi associated PDZ and coiled-coil motif containing isoform b | | Authors: | Li, X, Wu, J, Shi, Y. | | Deposit date: | 2005-12-20 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of GOPC PDZ domain and its interaction with the C-terminal motif of neuroligin
Protein Sci., 15, 2006
|
|
4HYG
 
 | | Structure of a presenilin family intramembrane aspartate protease in C222 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
2HNU
 
 | | Crystal Structure of a Dipeptide Complex of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2HNV
 
 | | Crystal Structure of a Dipeptide Complex of the Q58V Mutant of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
4HYC
 
 | | Structure of a presenilin family intramembrane aspartate protease in P2 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
2HNW
 
 | | Crystal Structure of the F91STOP mutant of des1-6 Bovine Neurophysin-I, unliganded state | | Descriptor: | Oxytocin-neurophysin 1 | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
7EQH
 
 | |
