4QY2
 
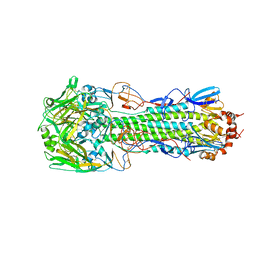 | | Structure of H10 from human-infecting H10N8 virus in complex with human receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4RFP
 
 | |
2G5T
 
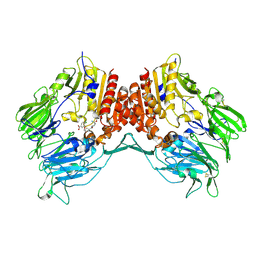 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) complexed with cyanopyrrolidine (C5-pro-pro) inhibitor 21ag | | Descriptor: | 3-{[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHOXY}-4-CHLOROBENZOIC ACID, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Fry, E.H, Lake, M.R, Solomon, L.R, Pei, Z, Li, X. | | Deposit date: | 2006-02-23 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery, structure-activity relationship, and pharmacological evaluation of (5-substituted-pyrrolidinyl-2-carbonyl)-2-cyanopyrrolidines as potent dipeptidyl peptidase IV inhibitors.
J.Med.Chem., 49, 2006
|
|
6BK8
 
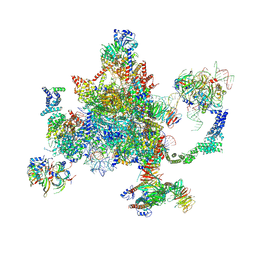 | | S. cerevisiae spliceosomal post-catalytic P complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Lea1, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2017-11-07 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the yeast spliceosomal postcatalytic P complex.
Science, 358, 2017
|
|
2GO4
 
 | | Crystal structure of Aquifex aeolicus LpxC complexed with TU-514 | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
2G63
 
 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) complexed with cyanopyrrolidine (C5-pro-pro) inhibitor 24b | | Descriptor: | Dipeptidyl peptidase 4, METHYL 4-{[({[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHYL}AMINO)CARBONYL]AMINO}BENZOATE | | Authors: | Longenecker, K.L, Fry, E.H, Lake, M.R, Solomon, L.R, Pei, Z, Li, X. | | Deposit date: | 2006-02-24 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery, structure-activity relationship, and pharmacological evaluation of (5-substituted-pyrrolidinyl-2-carbonyl)-2-cyanopyrrolidines as potent dipeptidyl peptidase IV inhibitors.
J.Med.Chem., 49, 2006
|
|
3HVN
 
 | | Crystal structure of cytotoxin protein suilysin from Streptococcus suis | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, HEPTANE-1,2,3-TRIOL, Hemolysin | | Authors: | Xu, L, Huang, B, Du, H, Zhang, C.X, Xu, J, Li, X, Rao, Z. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Crystal structure of cytotoxin protein suilysin from Streptococcus suis.
Protein Cell, 1, 2010
|
|
6C9U
 
 | | Crystal structure of [KS3][AT3] didomain from module 3 of 6-deoxyerthronolide B synthase in complex with antibody fragment (Fab) | | Descriptor: | 6-deoxyerythronolide-B synthase EryA2, modules 3 and 4, Heavy chain of Fab 1B2, ... | | Authors: | Deis, L.N, Li, X, Mathews, I.I, Khosla, C. | | Deposit date: | 2018-01-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Function Analysis of the Extended Conformation of a Polyketide Synthase Module.
J. Am. Chem. Soc., 140, 2018
|
|
4TVV
 
 | | Crystal structure of LppA from Legionella pneumophila | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Weber, S, Stirnimann, C, Wieser, M, Meier, R, Engelhardt, S, Li, X, Capitani, G, Kammerer, R, Hilbi, H. | | Deposit date: | 2014-06-28 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Type IV Translocated Legionella Cysteine Phytase Counteracts Intracellular Growth Restriction by Phytate.
J.Biol.Chem., 289, 2014
|
|
2G5P
 
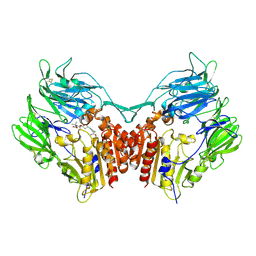 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) complexed with cyanopyrrolidine (C5-pro-pro) inhibitor 21ac | | Descriptor: | 4-{[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHOXY}-3-TERT-BUTYLBENZOIC ACID, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Fry, E.H, Lake, M.R, Solomon, L.R, Pei, Z, Li, X. | | Deposit date: | 2006-02-23 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery, structure-activity relationship, and pharmacological evaluation of (5-substituted-pyrrolidinyl-2-carbonyl)-2-cyanopyrrolidines as potent dipeptidyl peptidase IV inhibitors.
J.Med.Chem., 49, 2006
|
|
2K3C
 
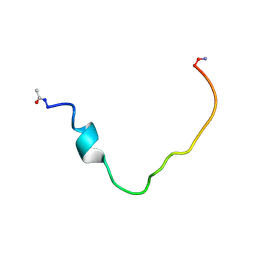 | | Structural and Functional Characterization of TM IX of the NHE1 Isoform of the Na+/H+ Exchanger | | Descriptor: | TMIX peptide | | Authors: | Reddy, T, Ding, J, Li, X, Sykes, B.D, Fliegel, L, Rainey, J.K. | | Deposit date: | 2008-05-01 | | Release date: | 2008-06-03 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Transmembrane Segment IX of the NHE1 Isoform of the Na+/H+ Exchanger.
J.Biol.Chem., 283, 2008
|
|
2GO3
 
 | | Crystal structure of Aquifex aeolicus LpxC complexed with imidazole. | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
6KJ3
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.60 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.6 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KG7
 
 | | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Piezo-type mechanosensitive ion channel component 2 | | Authors: | Wang, L, Zhou, H, Zhang, M, Liu, W, Deng, T, Zhao, Q, Li, Y, Lei, J, Li, X, Xiao, B. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanogating of the mammalian tactile channel PIEZO2.
Nature, 573, 2019
|
|
6E7P
 
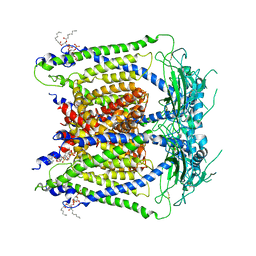 | | cryo-EM structure of human TRPML1 with PI35P2 | | Descriptor: | (1R,2S,3S,4R,5S,6R)-5-{[(R)-[(2R)-2,3-bis{[(1S)-1-hydroxyoctyl]oxy}propoxy](hydroxy)phosphoryl]oxy}-2,4,6-trihydroxycyclohexane-1,3-diyl bis[dihydrogen (phosphate)], Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
6E7Y
 
 | | cryo-EM structure of human TRPML1 with PI45P2 | | Descriptor: | Mucolipin-1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
6E7Z
 
 | | cryo-EM structure of human TRPML1 with ML-SA1 and PI35P2 | | Descriptor: | (1R,2S,3S,4R,5S,6R)-5-{[(R)-[(2R)-2,3-bis{[(1S)-1-hydroxyoctyl]oxy}propoxy](hydroxy)phosphoryl]oxy}-2,4,6-trihydroxycyclohexane-1,3-diyl bis[dihydrogen (phosphate)], 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
2I78
 
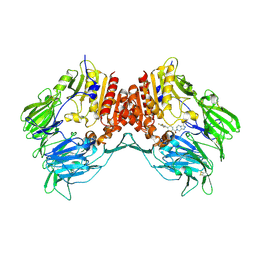 | | Crystal structure of human dipeptidyl peptidase IV (DPP IV) complexed with ABT-341, a cyclohexene-constrained phenethylamine inhibitor | | Descriptor: | (1S,6R)-3-{[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]CARBONYL}-6-(2,4,5-TRIFLUOROPHENYL)CYCLOHEX-3-EN-1-AMINE, Dipeptidyl peptidase IV | | Authors: | Longenecker, K.L, Pei, Z, Li, X. | | Deposit date: | 2006-08-30 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Cyclohexene-constrained Phenethylamine ABT-341, a Highly Potent, Selective, Orally Bioavailable, Safe and Potential Next-generation Dipeptidyl Peptidase IV Inhibitor for the Treatment of Type 2 Diabetes
To be Published
|
|
1C9W
 
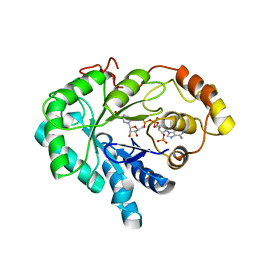 | | CHO REDUCTASE WITH NADP+ | | Descriptor: | CHO REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Li, X, Hyndman, D, Flynn, T.G, Jia, Z. | | Deposit date: | 1999-08-03 | | Release date: | 2000-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
Proteins, 38, 2000
|
|
2GVE
 
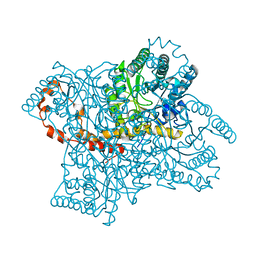 | | Time-of-Flight Neutron Diffraction Structure of D-Xylose Isomerase | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Li, X, Carrell, H.L, Hanson, B.L, Langan, P, Coates, L, Schoenborn, B.P, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-05-02 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8Z0K
 
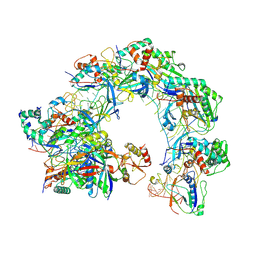 | | Cryo-EM structure of Cas8-HNH system at full R-loop state | | Descriptor: | DNA (37-MER), DNA (5'-D(P*GP*TP*GP*CP*GP*GP*A)-3'), HNH endonuclease, ... | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-04-09 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
5CMW
 
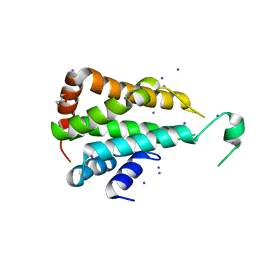 | | Crystal structure of yeast Ent5 N-terminal domain-soaked in KI | | Descriptor: | Epsin-5, GLYCEROL, IODIDE ION | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Biochem.Biophys.Res.Commun., 477, 2016
|
|
8YJC
 
 | | Structure of Vibrio vulnificus MARTX cysteine protease domain C3727A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, INOSITOL HEXAKISPHOSPHATE, Multifunctional autoprocessing repeat-in-toxin (MARTX), ... | | Authors: | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | Deposit date: | 2024-03-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of the activation of MARTX cysteine protease domain from Vibrio vulnificus.
Plos One, 19, 2024
|
|
8ZNR
 
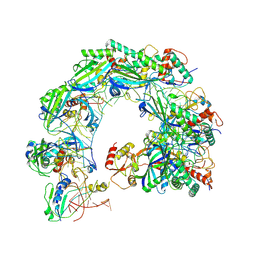 | | Cryo-EM structure of Cas8-HNH system at ssDNA-bound state | | Descriptor: | DNA (35-MER), RNA (56-MER), protein structure | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
