1XSP
 
 | | Crystal Structure of human DNA polymerase lambda in complex with nicked DNA and pyrophosphate | | Descriptor: | 5'-D(*CP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*TP*AP*CP*TP*G)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1XSN
 
 | | Crystal Structure of human DNA polymerase lambda in complex with a one nucleotide DNA gap and ddTTP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*(2DT))-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
3UAN
 
 | | Crystal structure of 3-O-sulfotransferase (3-OST-1) with bound PAP and heptasaccharide substrate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Moon, A.F, Xu, Y, Woody, S.M, Krahn, J.M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7TQV
 
 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (33-MER), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
7TJ2
 
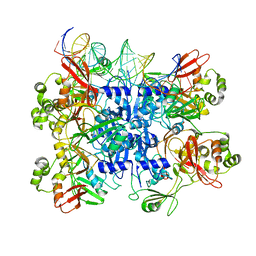 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (31-MER), Uridylate-specific endoribonuclease nsp15 | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
8V5D
 
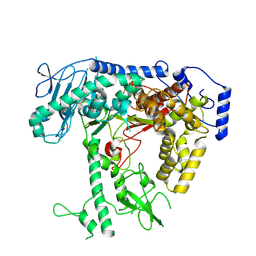 | | Human mitochondrial DNA polymerase catalytic subunit, PolG, in an APO conformation | | Descriptor: | DNA polymerase subunit gamma-1 | | Authors: | Riccio, A.A, Brannon, A.J, Krahn, J.M, Bouvette, J, Borgnia, J.M, Copeland, W.C. | | Deposit date: | 2023-11-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Coordinated DNA polymerization by Pol gamma and the region of LonP1 regulated proteolysis.
Nucleic Acids Res., 52, 2024
|
|
8V54
 
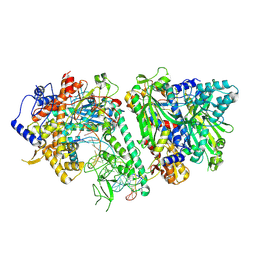 | | Engaged conformation of the human mitochondrial DNA polymerase gamma bound to DNA | | Descriptor: | DNA (25-MER), DNA (44-MER), DNA polymerase subunit gamma-1, ... | | Authors: | Riccio, A.A, Krahn, J.M, Bouvette, J, Borgnia, J.M, Copeland, W.C. | | Deposit date: | 2023-11-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Coordinated DNA polymerization by Pol gamma and the region of LonP1 regulated proteolysis.
Nucleic Acids Res., 52, 2024
|
|
8V5R
 
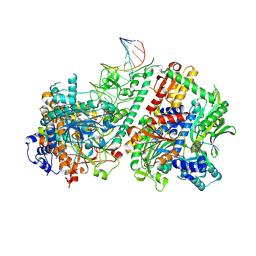 | | Active conformation of DNA polymerase gamma bound to DNA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, ... | | Authors: | Riccio, A.A, Krahn, J.M, Bouvette, J, Borgnia, J.M, Copeland, W.C. | | Deposit date: | 2023-11-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Coordinated DNA polymerization by Pol gamma and the region of LonP1 regulated proteolysis.
Nucleic Acids Res., 52, 2024
|
|
7SCD
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-1 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7SCE
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-2 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7N33
 
 | | SARS-CoV-2 Nsp15 endoribonuclease pre-cleavage state | | Descriptor: | RNA (5'-R(*A)-D(*(UFT))-R(P*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
7N06
 
 | | SARS-CoV-2 Nsp15 endoribonuclease post-cleavage state | | Descriptor: | RNA (5'-R(*AP*UP*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
6OF3
 
 | | Precursor ribosomal RNA processing complex, State 1. | | Descriptor: | CLP1_P domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OF2
 
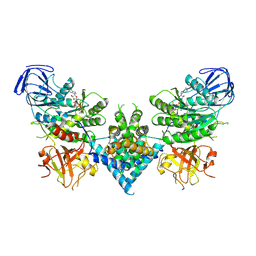 | | Precursor ribosomal RNA processing complex, State 2. | | Descriptor: | CLP1_P domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OF4
 
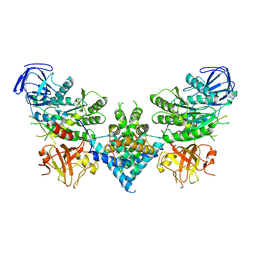 | | Precursor ribosomal RNA processing complex, apo-state. | | Descriptor: | CLP1_P domain-containing protein, Ribonuclease | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q00
 
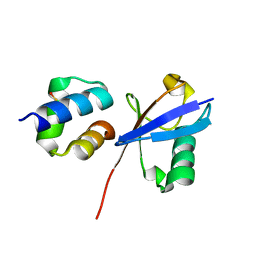 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 1 | | Descriptor: | POTASSIUM ION, Tyrosyl-DNA phosphodiesterase 2, Ubiquitin | | Authors: | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-29 | | Last modified: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
6Q01
 
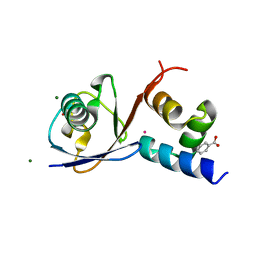 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 2 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.851 Å) | | Cite: | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
1ECJ
 
 | |
4IM8
 
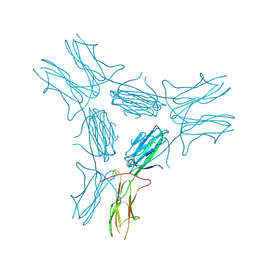 | | low resolution crystal structure of mouse RAGE | | Descriptor: | Advanced glycation end-products receptor | | Authors: | Xu, D, Young, J.H, Krahn, J.M, Song, D, Corbett, K.D, Chazin, W.J, Pedersen, L.C, Esko, J.D. | | Deposit date: | 2013-01-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Stable RAGE-Heparan Sulfate Complexes Are Essential for Signal Transduction.
Acs Chem.Biol., 8, 2013
|
|
2FMS
 
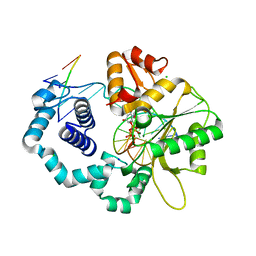 | | DNA Polymerase beta with a gapped DNA substrate and dUMPNPP with magnesium in the catalytic site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
2FMQ
 
 | | Sodium in active site of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
2FMP
 
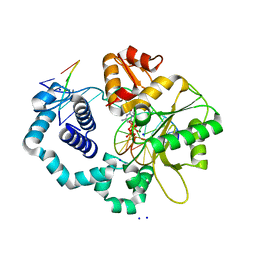 | | DNA Polymerase beta with a terminated gapped DNA substrate and ddCTP with sodium in the catalytic site | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
4KSE
 
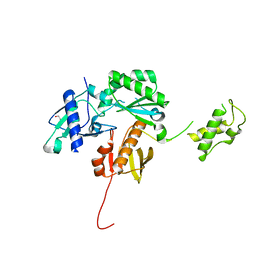 | | Crystal structure of a HIV p51 (219-230) deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, HIV p51 subunit | | Authors: | Zheng, X, Mueller, G.A, Derose, E.F, Pedersen, L.C, Gabel, S.A, Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2013-05-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation.
Nucleic Acids Res., 42, 2014
|
|
8V55
 
 | | Human mitochondrial DNA polymerase gamma bound to a replication fork in an open conformation | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Riccio, A.A, Krahn, J.M, Bouvette, J, Borgnia, M.J, Copeland, W.C. | | Deposit date: | 2023-11-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Coordinated DNA polymerization by Pol gamma and the region of LonP1 regulated proteolysis.
Nucleic Acids Res., 52, 2024
|
|
8V58
 
 | | Complex of murine cathepsin K with bound heparan sulfate 12mer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Pedersen, L.C, Xu, D, Krahn, J.M. | | Deposit date: | 2023-11-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Heparan sulfate selectively inhibits the collagenase activity of cathepsin K.
Matrix Biol., 129, 2024
|
|
