4N9X
 
 | | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161 | | Descriptor: | Putative monooxygenase | | Authors: | Kuzin, A, Chen, Y, Lew, S, Seetharaman, J, Mao, L, Xiao, R, Owens, L.A, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-21 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161
To be Published
|
|
4N4Y
 
 | |
3UYI
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
4Q2V
 
 | | Crystal Structure of Ricin A chain complexed with Baicalin inhibitor | | Descriptor: | 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, Ricin | | Authors: | Deng, X, Li, X, Dong, J, Chen, Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Baicalin inhibits the lethality of ricin in mice by inducing protein oligomerization.
J.Biol.Chem., 290, 2015
|
|
8JOT
 
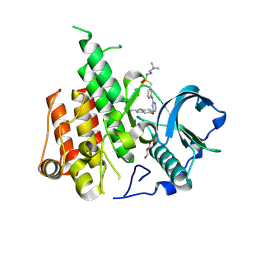 | | Crystal structure of CSF-1R kinase domain with sulfatinib | | Descriptor: | GLYCEROL, Macrophage colony-stimulating factor 1 receptor, N-(2-(dimethylamino)ethyl)-1-(3-((4-((2-methyl-1H-indol-5-yl)oxy)pyrimidin-2-yl)amino)phenyl)methanesulfonamide | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
8JMZ
 
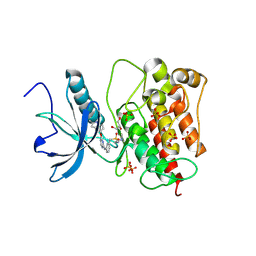 | | FGFR1 kinase domain with sulfatinib | | Descriptor: | Fibroblast growth factor receptor 1, GLYCEROL, N-(2-(dimethylamino)ethyl)-1-(3-((4-((2-methyl-1H-indol-5-yl)oxy)pyrimidin-2-yl)amino)phenyl)methanesulfonamide, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2023-06-05 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
5WFI
 
 | |
4FKZ
 
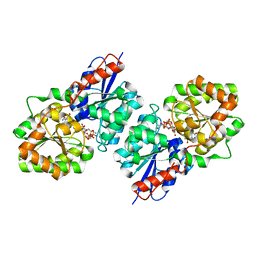 | | Crystal structure of Bacillus subtilis UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP | | Descriptor: | UDP-N-acetylglucosamine 2-epimerase, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Yang, C.S, Chen, S.C, Kuan, S.M, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2012-06-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of Bacillus subtilis UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP
To be Published
|
|
4PAS
 
 | | Heterodimeric coiled-coil structure of human GABA(B) receptor | | Descriptor: | Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Burmakina, S, Geng, Y, Chen, Y, Fan, Q.R. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Heterodimeric coiled-coil interactions of human GABAB receptor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8JSF
 
 | |
8JSJ
 
 | |
8JSK
 
 | | Crystal structure of an N-terminal cyclic nucleotide-binding domain of a PycTIR from Pseudovibrio sp. in complex with cUMP | | Descriptor: | PycTIR, Uridine-3',5'-cyclic monophosphate | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-06-20 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural and functional characterization of cyclic pyrimidine-regulated anti-phage system.
Nat Commun, 15, 2024
|
|
8JSZ
 
 | | Crystal structure of a uridylate cyclase from Anabaena sp. | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-06-20 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural and functional characterization of cyclic pyrimidine-regulated anti-phage system.
Nat Commun, 15, 2024
|
|
7VUE
 
 | | Structural insight of the molecular mechanism of cilofexor bound to FXR | | Descriptor: | 2-[3-[4-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-2-chloranyl-phenyl]-3-oxidanyl-azetidin-1-yl]pyridine-4-carboxylic acid, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Jiang, L, Chen, Y.C. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural insight into the molecular mechanism of cilofexor binding to the farnesoid X receptor.
Biochem.Biophys.Res.Commun., 595, 2022
|
|
5VLE
 
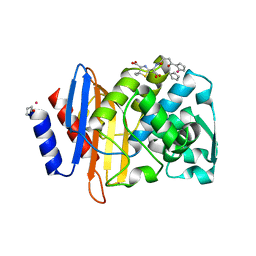 | |
5WQD
 
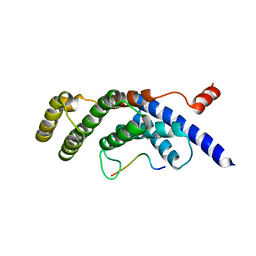 | |
4Q9V
 
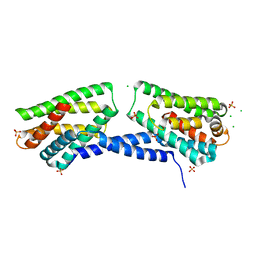 | | Crystal structure of TIPE3 | | Descriptor: | CHLORIDE ION, SULFATE ION, Tumor necrosis factor alpha-induced protein 8-like protein 3 | | Authors: | Wu, J, Zhang, X, Chen, Y.H, Shi, Y. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Cancer Cell, 26, 2014
|
|
4QLA
 
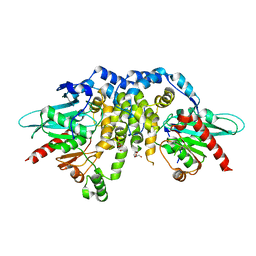 | | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori | | Descriptor: | Juvenile hormone epoxide hydrolase, PENTAETHYLENE GLYCOL | | Authors: | Zhou, K, Jia, N, Hu, C, Jiang, Y.L, Yang, J.P, Chen, Y, Li, S, Zhou, C.Z. | | Deposit date: | 2014-06-11 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori.
Proteins, 82, 2014
|
|
3U4T
 
 | | Crystal Structure of the C-terminal part of the TPR repeat-containing protein Q11TI6_CYTH3 from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target ChR11B. | | Descriptor: | TPR repeat-containing protein | | Authors: | Vorobiev, S, Neely, H, Chen, Y, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-10 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Crystal Structure of the C-terminal part of the TPR repeat-containing protein Q11TI6_CYTH3 from Cytophaga hutchinsonii.
To be Published
|
|
8D4K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4M
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
6A3N
 
 | | Crystal structure of the PDE9 catalytic domain in complex with inhibitor 2 | | Descriptor: | 1-cyclopentyl-6-({(2R)-1-[(3S)-3-fluoropyrrolidin-1-yl]-1-oxopropan-2-yl}amino)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wu, Y.N, Zhou, Q, Chen, Y.P, Luo, H.B. | | Deposit date: | 2018-06-15 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors against Phosphodiesterase-9, a Novel Target for the Treatment of Vascular Dementia.
J. Med. Chem., 62, 2019
|
|
3HKS
 
 | | Crystal structure of eukaryotic translation initiation factor eIF-5A2 from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, Eukaryotic translation initiation factor 5A-2 | | Authors: | Teng, Y.B, He, Y.X, Jiang, Y.L, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2009-05-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Arabidopsis translation initiation factor eIF-5A2
Proteins, 77, 2009
|
|
3V0T
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine Reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.333 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3V0S
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
