8T7C
 
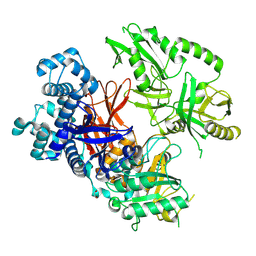 | | Crystal structure of human phospholipase C gamma 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2, CALCIUM ION | | Authors: | Chen, Y, Choi, H, Zhuang, N, Hu, L, Qian, D, Wang, J. | | Deposit date: | 2023-06-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal and cryo-EM structures of PLCg2 reveal dynamic inter-domain recognitions in autoinhibition
To Be Published
|
|
8GV7
 
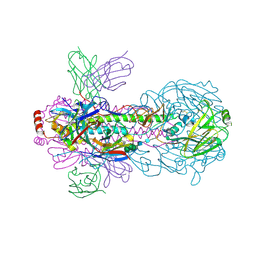 | | Crystal structure of PN-SIA28 in complex with influenza hemagglutinin H18 A/flat-faced bat/Peru/033/2010 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin H18-HA1, ... | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
8GK7
 
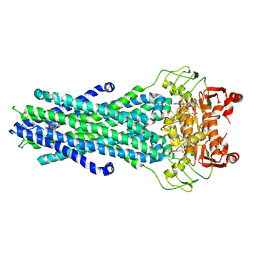 | | MsbA bound to cerastecin C | | Descriptor: | 2-[(4-butylbenzene-1-sulfonyl)amino]-5-[(3-{4-[(4-butylbenzene-1-sulfonyl)amino]-3-carboxyanilino}-3-oxopropyl)carbamoyl]benzoic acid, Lipid A export ATP-binding/permease protein MsbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Y, Klein, D. | | Deposit date: | 2023-03-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cerastecins inhibit membrane lipooligosaccharide transport in drug-resistant Acinetobacter baumannii.
Nat Microbiol, 9, 2024
|
|
7EQT
 
 | |
7EQS
 
 | |
7EQW
 
 | |
7ER1
 
 | |
7ER0
 
 | |
8GV5
 
 | | Crystal structure of PN-SIA28 in complex with influenza hemagglutinin A/swine/Guangdong/104/2013 (H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
7FJD
 
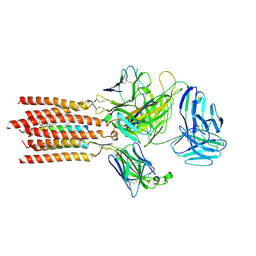 | | Cryo-EM structure of a membrane protein(WT) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJE
 
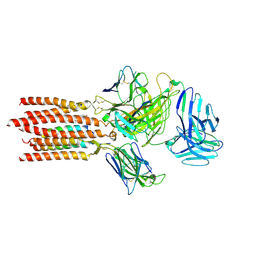 | | Cryo-EM structure of a membrane protein(LL) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJF
 
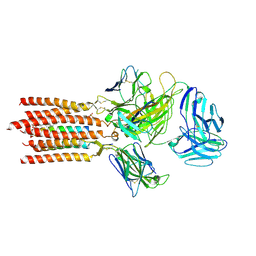 | | Cryo-EM structure of a membrane protein(CS) | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
8KGO
 
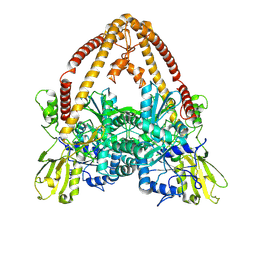 | |
8GV6
 
 | | Crystal structure of PN-SIA28 in complex with influenza hemagglutinin H14 (A/long-tailed duck/Wisconsin/10OS3912/2010) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin H14-HA1, ... | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
8GV4
 
 | | A human broadly neutralizing influenza A hemagglutinin stem-specific antibody PN-SIA28 | | Descriptor: | PN-SIA28 heavy chain, PN-SIA28 light chain | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
7DDC
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with Tafenoquine | | Descriptor: | 3C-like proteinase, Tafenoquine | | Authors: | Chen, Y, Wang, Y.C, Yang, C.S, Hung, M.C. | | Deposit date: | 2020-10-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Tafenoquine and its derivatives as inhibitors for the severe acute respiratory syndrome coronavirus 2.
J.Biol.Chem., 298, 2022
|
|
6KHY
 
 | | The crystal structure of AsfvAP:AG | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (AGCGTCACCGACGAGGC), DNA(AGCGTCACCGACGAGG), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
2HKO
 
 | | Crystal structure of LSD1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1 | | Authors: | Chen, Y, Yang, Y.T, Wang, F, Yanane, K, Zhang, Y, Lei, M. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human histone lysine-specific demethylase 1 (LSD1).
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8CTM
 
 | |
7X4A
 
 | | Native CD-NTase ClCdnE | | Descriptor: | ClCdnE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4G
 
 | | CD-NTase ClCdnE in complex with substrate UTP | | Descriptor: | ClCdnE, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
8CZZ
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with Temsavir, 8ANC195, and 10-1074 | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Pozharski, E, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
7UL1
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the neutralizing IGHV3-53-encoded antibody EH3 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH3, Light chain of EH3, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UL0
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the ridge-binding nAb EH8 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH8, ... | | Authors: | Chen, Y, Tolbert, W.D, Bai, X, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
8DOK
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
