5XP7
 
 | | C-Src in complex with ATP-CHCl | | Descriptor: | GLYCEROL, MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, ... | | Authors: | Guo, M, Dai, S, Duan, Y, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
7S0V
 
 | |
3UCI
 
 | | Crystal structure of Rhodostomin ARLDDL mutant | | Descriptor: | disintegrin | | Authors: | Shiu, J.H, Chen, C.Y, Chen, Y.C, Chang, Y.T, Chang, Y.S, Huang, C.H, Chuang, W.J. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
6CSU
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 152 kDa, Centrosomal protein of 63 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
2XR0
 
 | | Room temperature X-ray structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | SULFATE ION, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
5YI6
 
 | | CRISPR associated protein Cas6 | | Descriptor: | CRISPR-associated endoribonuclease Cas6 1, GLYCEROL, PHOSPHATE ION | | Authors: | Ko, T.P, Hsieh, T.J, Chen, Y. | | Deposit date: | 2017-10-03 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Expression, Purification, Crystallization, and X-ray Structural Analysis of CRISPR-Associated Protein Cas6 from Methanocaldococcus jannaschii
Crystals, 7, 2018
|
|
4E12
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
4E8C
 
 | | Crystal structure of streptococcal beta-galactosidase in complex with galactose | | Descriptor: | GLYCEROL, Glycosyl hydrolase, family 35, ... | | Authors: | Cheng, W, Wang, L, Bai, X.H, Jiang, Y.L, Li, Q, Yu, G, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
J.Biol.Chem., 287, 2012
|
|
4E8D
 
 | | Crystal structure of streptococcal beta-galactosidase | | Descriptor: | GLYCEROL, Glycosyl hydrolase, family 35 | | Authors: | Cheng, W, Wang, L, Bai, X.H, Jiang, Y.L, Li, Q, Yu, G, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
J.Biol.Chem., 287, 2012
|
|
4HMO
 
 | | Crystal structure of streptococcus pneumoniae TIGR4 PiaA in complex with Bis-tris propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Iron-compound ABC transporter, ... | | Authors: | Cheng, W, Li, Q, Jiang, Y.-L, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2012-10-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Streptococcus pneumoniae PiaA and Its Complex with Ferrichrome Reveal Insights into the Substrate Binding and Release of High Affinity Iron Transporters
Plos One, 8, 2013
|
|
6Q2N
 
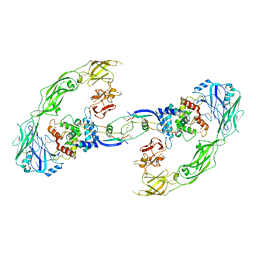 | | Cryo-EM structure of RET/GFRa1/GDNF extracellular complex | | Descriptor: | CALCIUM ION, GDNF family receptor alpha-1, Glial cell line-derived neurotrophic factor, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
1MN9
 
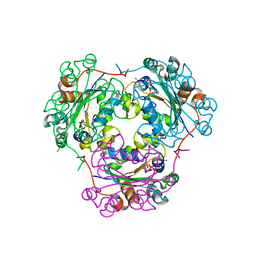 | | NDP kinase mutant (H122G) complex with RTP | | Descriptor: | MAGNESIUM ION, NDP kinase, RIBAVIRIN TRIPHOSPHATE | | Authors: | Gallois-montbrun, S, Chen, Y, Dutartre, H, Morera, S, Guerreiro, C, Mulard, L, Schneider, B, Janin, J, Canard, B, Veron, M, Deville-bonne, D. | | Deposit date: | 2002-09-05 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Analysis of the Activation of Ribavirin Analogs by NDP Kinase: Comparison with Other Ribavirin Targets
MOL.PHARMACOL., 63, 2003
|
|
3B5D
 
 | |
1B6C
 
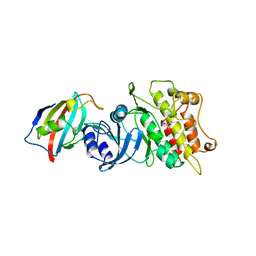 | | CRYSTAL STRUCTURE OF THE CYTOPLASMIC DOMAIN OF THE TYPE I TGF-BETA RECEPTOR IN COMPLEX WITH FKBP12 | | Descriptor: | FK506-BINDING PROTEIN, SULFATE ION, TGF-B SUPERFAMILY RECEPTOR TYPE I | | Authors: | Huse, M, Chen, Y.-G, Massague, J, Kuriyan, J. | | Deposit date: | 1999-01-13 | | Release date: | 1999-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the cytoplasmic domain of the type I TGF beta receptor in complex with FKBP12.
Cell(Cambridge,Mass.), 96, 1999
|
|
1IAY
 
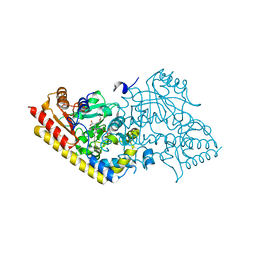 | | CRYSTAL STRUCTURE OF ACC SYNTHASE COMPLEXED WITH COFACTOR PLP AND INHIBITOR AVG | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2, 2-AMINO-4-(2-AMINO-ETHOXY)-BUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Huai, Q, Xia, Y, Chen, Y, Callahan, B, Li, N, Ke, H. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5'-phosphate provide new insight into catalytic mechanisms
J.Biol.Chem., 276, 2001
|
|
4L2Y
 
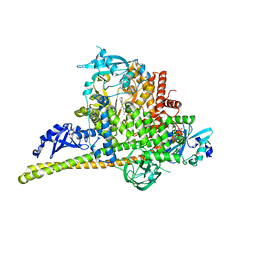 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and compound 9d | | Descriptor: | 3-amino-5-[4-(morpholin-4-yl)pyrido[3',2':4,5]furo[3,2-d]pyrimidin-2-yl]phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
4L1B
 
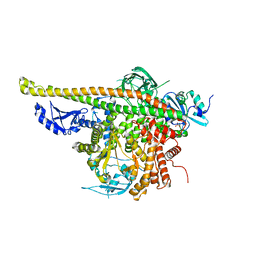 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
3B61
 
 | |
3B62
 
 | |
4L23
 
 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and PI-103 | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
1XTO
 
 | | Crystal Structure of the Coenzyme PQQ Synthesis Protein (PqqB) from Pseudomonas putida, Northeast Structural Genomics Target PpR6 | | Descriptor: | Coenzyme PQQ synthesis protein B, ZINC ION | | Authors: | Forouhar, F, Chen, Y, Kuzin, A, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-22 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Coenzyme PQQ Synthesis Protein (PqqB) from Pseudomonas putida, Northeast Structural Genomics Target PpR6
To be Published
|
|
4D6V
 
 | | Crystal structure of signal transducing protein | | Descriptor: | G PROTEIN BETA SUBUNIT GIB2 | | Authors: | Ero, R, Dimitrova, V.T, Chen, Y, Bu, W, Feng, S, Liu, T, Wang, P, Xue, C, Tan, S.M, Gao, Y.G. | | Deposit date: | 2014-11-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Gib2, a Signal-Transducing Protein Scaffold Associated with Ribosomes in Cryptococcus Neoformans.
Sci.Rep., 5, 2015
|
|
3EH4
 
 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
1XW8
 
 | | X-ray structure of putative lactam utilization protein YBGL. Northeast Structural Genomics Consortium target ET90. | | Descriptor: | UPF0271 protein ybgL | | Authors: | Kuzin, A.P, Chen, Y, Vorobiev, S.M, Acton, T.B, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of putative lactam utilization protein YBGL.
Northeast Structural Genomics Consortium target ET90.
To be Published
|
|
