7RPK
 
 | | Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog | | 分子名称: | (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | 登録日 | 2021-08-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPJ
 
 | | Cryo-EM structure of murine Dispatched NNN mutant | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | 著者 | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | 登録日 | 2021-08-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2021-11-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPH
 
 | | Cryo-EM structure of murine Dispatched 'R' conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | 著者 | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | 登録日 | 2021-08-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPI
 
 | | Cryo-EM structure of murine Dispatched 'T' conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | 著者 | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | 登録日 | 2021-08-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
6JVJ
 
 | | Crystal structure of human MTH1 in complex with compound MI1006 | | 分子名称: | 5-ethyl-N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase | | 著者 | Peng, C, Li, Y.H, Cheng, Y.S. | | 登録日 | 2019-04-17 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.297 Å) | | 主引用文献 | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVR
 
 | | Crystal structure of human MTH1 in complex with compound MI1026 | | 分子名称: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | 著者 | Peng, C, Li, Y.H, Cheng, Y.S. | | 登録日 | 2019-04-17 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
8U3L
 
 | |
8U30
 
 | | TRPV1 in nanodisc bound with diC8-PIP2 in the closed state | | 分子名称: | SODIUM ION, Transient receptor potential cation channel subfamily V member 1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | 著者 | Arnold, W.R, Julius, D, Cheng, Y. | | 登録日 | 2023-09-06 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8U2Z
 
 | | TRPV1 in nanodisc bound with diC8-PIP2 in the dilated state | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, SODIUM ION, ... | | 著者 | Arnold, W.R, Julius, D, Cheng, Y. | | 登録日 | 2023-09-06 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8U3C
 
 | | TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 2 (monomer) | | 分子名称: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Arnold, W.R, Julius, D, Cheng, Y. | | 登録日 | 2023-09-07 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8U3J
 
 | |
8U4D
 
 | | TRPV1 in nanodisc bound with PI-Br4, consensus structure | | 分子名称: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | 著者 | Arnold, W.R, Julius, D, Cheng, Y. | | 登録日 | 2023-09-10 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8U3A
 
 | | TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 1 (monomer) | | 分子名称: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, SODIUM ION, ... | | 著者 | Arnold, W.R, Julius, D, Cheng, Y. | | 登録日 | 2023-09-07 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8U43
 
 | | TRPV1 in nanodisc bound with PIP2-Br4 | | 分子名称: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, SODIUM ION, ... | | 著者 | Arnold, W.R, Julius, D, Cheng, Y. | | 登録日 | 2023-09-08 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
6JVI
 
 | | Crystal structure of human MTH1 in complex with compound MI0861 | | 分子名称: | (4R)-4-(2-methoxyphenyl)-4,6,7,8-tetrahydroquinoline-2,5(1H,3H)-dione, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | 著者 | Peng, C, Cheng, Y.S. | | 登録日 | 2019-04-17 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.249 Å) | | 主引用文献 | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVP
 
 | | Crystal structure of human MTH1 in complex with compound MI1024 | | 分子名称: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | 著者 | Peng, C, Li, Y.H, Cheng, Y.S. | | 登録日 | 2019-04-17 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVG
 
 | | Crystal structure of human MTH1 in complex with compound MI0639 | | 分子名称: | 5-ethyl-4-methyl-6-(morpholin-4-yl)pyrimidin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase | | 著者 | Peng, C, Cheng, Y.S. | | 登録日 | 2019-04-17 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.844 Å) | | 主引用文献 | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVQ
 
 | | Crystal structure of human MTH1 in complex with compound MI1025 | | 分子名称: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-morpholin-4-yl-pyrimidine-2,4-diamine | | 著者 | Peng, C, Li, Y.H, Cheng, Y.S. | | 登録日 | 2019-04-17 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.197 Å) | | 主引用文献 | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVO
 
 | | Crystal structure of human MTH1 in complex with compound MI1022 | | 分子名称: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-(4-methylpiperazin-1-yl)pyrimidine-2,4-diamine | | 著者 | Peng, C, Li, Y.H, Cheng, Y.S. | | 登録日 | 2019-04-17 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVH
 
 | |
3AMS
 
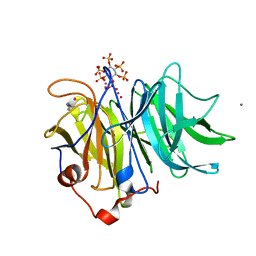 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Cd2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | 分子名称: | 3-phytase, CADMIUM ION, CALCIUM ION, ... | | 著者 | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | 登録日 | 2010-08-22 | | 公開日 | 2011-04-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
8V4Y
 
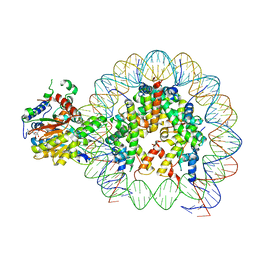 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Histone H2A type 1, ... | | 著者 | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | 登録日 | 2023-11-29 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8V7L
 
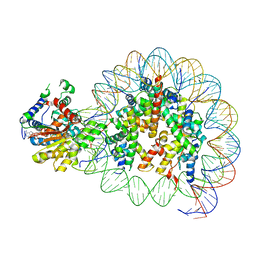 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | 著者 | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | 登録日 | 2023-12-04 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8V6V
 
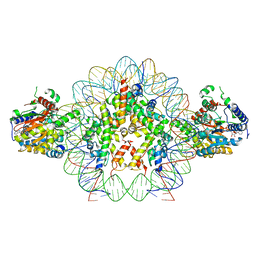 | | Cryo-EM structure of doubly-bound SNF2h-nucleosome complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | 著者 | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | 登録日 | 2023-12-03 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
6NE3
 
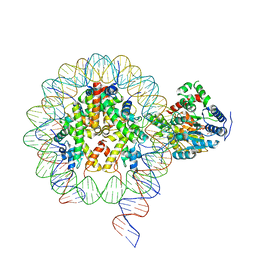 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h bound at SHL-2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (156-MER), Histone H2A type 1, ... | | 著者 | Armache, J.-P, Gamarra, N, Johnson, S.L, Leonard, J.D, Wu, S, Narlikar, G.N, Cheng, Y. | | 登録日 | 2018-12-16 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome.
Elife, 8, 2019
|
|
