3PB7
 
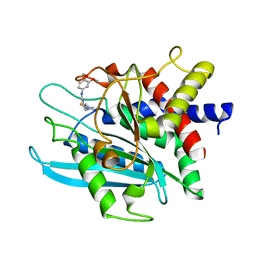 | | Crystal structure of the catalytic domain of human Golgi-resident glutaminyl cyclase in complex with PBD150 | | 分子名称: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, Glutaminyl-peptide cyclotransferase-like protein, ZINC ION | | 著者 | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | 登録日 | 2010-10-20 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
3PB9
 
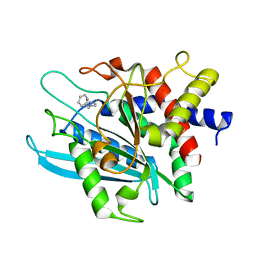 | | Crystal structure of the catalytic domain of human Golgi-resident glutaminyl cyclase in complex with 1-benzylimidazole | | 分子名称: | 1-BENZYL-1H-IMIDAZOLE, Glutaminyl-peptide cyclotransferase-like protein, ZINC ION | | 著者 | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | 登録日 | 2010-10-20 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
3PPO
 
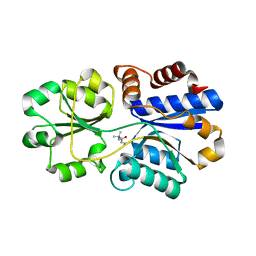 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | 分子名称: | (2S)-3-carboxy-2-hydroxy-N,N,N-trimethylpropan-1-aminium, Glycine betaine/carnitine/choline-binding protein | | 著者 | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | 登録日 | 2010-11-24 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
7X3O
 
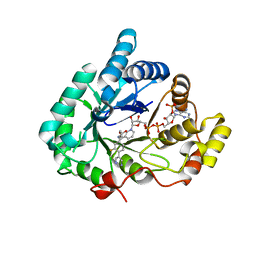 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054 | | 分子名称: | (2~{R})-2-(3-fluoranyl-4-pyrimidin-5-yl-phenyl)butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | 登録日 | 2022-03-01 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054
To Be Published
|
|
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | 分子名称: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | 登録日 | 2022-03-01 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | 分子名称: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | 登録日 | 2022-03-01 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.694 Å) | | 主引用文献 | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
3PBE
 
 | | Crystal structure of the mutant W207F of human secretory glutaminyl cyclase | | 分子名称: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | 著者 | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | 登録日 | 2010-10-20 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
3PPP
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | 分子名称: | Glycine betaine/carnitine/choline-binding protein, TRIMETHYL GLYCINE | | 著者 | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | 登録日 | 2010-11-25 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPN
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | 分子名称: | Glycine betaine/carnitine/choline-binding protein | | 著者 | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | 登録日 | 2010-11-24 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
7XY7
 
 | | Adenosine receptor bound to a non-selective agonist in complex with a G protein obtained by cryo-EM | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | 登録日 | 2022-05-31 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
7XY6
 
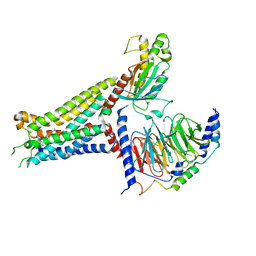 | | Adenosine receptor bound to an agonist in complex with G protein obtained by cryo-EM | | 分子名称: | 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2-yl]sulfanylethanamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | 登録日 | 2022-05-31 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
3PB8
 
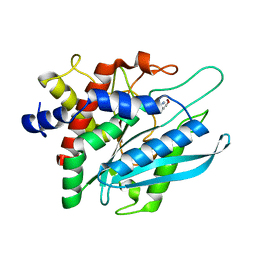 | | Crystal structure of the catalytic domain of human Golgi-resident glutaminyl cyclase in complex with N-acetylhistamine | | 分子名称: | Glutaminyl-peptide cyclotransferase-like protein, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE, ZINC ION | | 著者 | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | 登録日 | 2010-10-20 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
3QWB
 
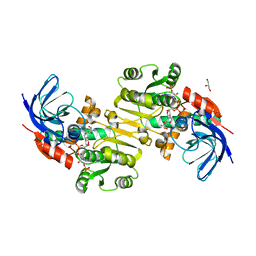 | | Crystal structure of Saccharomyces cerevisiae Zeta-crystallin-like quinone oxidoreductase Zta1 complexed with NADPH | | 分子名称: | GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable quinone oxidoreductase | | 著者 | Guo, P.C, Ma, X.X, Bao, Z.Z, Chen, Y.X, Zhou, C.Z. | | 登録日 | 2011-02-28 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1
J.Struct.Biol., 176, 2011
|
|
3QWA
 
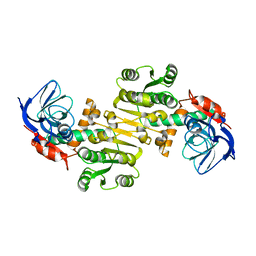 | | Crystal structure of Saccharomyces cerevisiae Zeta-crystallin-like quinone oxidoreductase Zta1 | | 分子名称: | Probable quinone oxidoreductase | | 著者 | Guo, P.C, Ma, X.X, Bao, Z.Z, Chen, Y.X, Zhou, C.Z. | | 登録日 | 2011-02-27 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1
J.Struct.Biol., 176, 2011
|
|
3QV0
 
 | | Crystal structure of Saccharomyces cerevisiae Mam33 | | 分子名称: | Mitochondrial acidic protein MAM33 | | 著者 | Jiang, Y.L, Pu, Y.G, Ma, X.X, Chen, Y, Zhou, C.Z. | | 登録日 | 2011-02-24 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures and putative interface of Saccharomyces cerevisiae mitochondrial matrix proteins Mmf1 and Mam33.
J.Struct.Biol., 175, 2011
|
|
3PPQ
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | 分子名称: | CHOLINE ION, Glycine betaine/carnitine/choline-binding protein | | 著者 | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | 登録日 | 2010-11-25 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
7XBK
 
 | | Structure and mechanism of a mitochondrial AAA+ disaggregase CLPB | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Isoform 2 of Caseinolytic peptidase B protein homolog, MAGNESIUM ION, ... | | 著者 | Wu, D, Liu, Y, Dai, Y, Wang, G, Lu, G, Chen, Y, Li, N, Lin, J, Gao, N. | | 登録日 | 2022-03-21 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
3R1G
 
 | | Structure Basis of Allosteric Inhibition of BACE1 by an Exosite-Binding Antibody | | 分子名称: | Beta-secretase 1, FAB of YW412.8.31 antibody heavy chain, FAB of YW412.8.31 antibody light chain | | 著者 | Wang, W, Rouge, L, Wu, P, Chiu, C, Chen, Y, Wu, Y, Watts, R.J. | | 登録日 | 2011-03-10 | | 公開日 | 2011-06-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Therapeutic Antibody Targeting BACE1 Inhibits Amyloid-{beta} Production in Vivo.
Sci Transl Med, 3, 2011
|
|
3PUB
 
 | | Crystal structure of the Bombyx mori low molecular weight lipoprotein 7 (Bmlp7) | | 分子名称: | 30kDa protein | | 著者 | Yang, J.-P, Ma, X.-X, He, Y.-X, Li, W.-F, Kang, Y, Bao, R, Chen, Y, Zhou, C.-Z. | | 登録日 | 2010-12-03 | | 公開日 | 2011-06-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Crystal structure of the 30 K protein from the silkworm Bombyx mori reveals a new member of the beta-trefoil superfamily
J.Struct.Biol., 175, 2011
|
|
3PPR
 
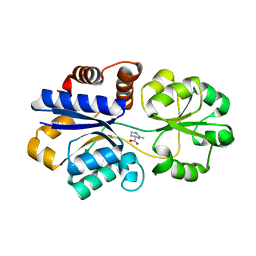 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | 分子名称: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Glycine betaine/carnitine/choline-binding protein | | 著者 | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | 登録日 | 2010-11-25 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
6O72
 
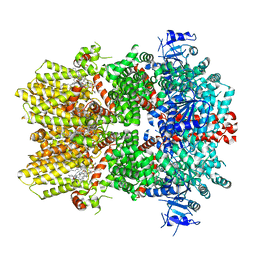 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, TC-I 2014-bound state | | 分子名称: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Diver, M.M, Cheng, Y, Julius, D. | | 登録日 | 2019-03-07 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
3CER
 
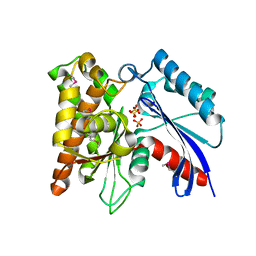 | | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13 | | 分子名称: | Possible exopolyphosphatase-like protein, SULFATE ION | | 著者 | Kuzin, A.P, Su, M, Chen, Y, Neely, H, Seetharaman, J, Shastry, R, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-02-29 | | 公開日 | 2008-04-01 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13.
To be Published
|
|
3P16
 
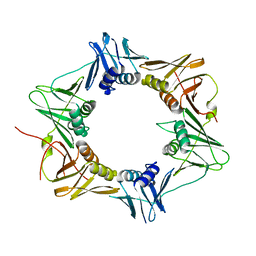 | | Crystal structure of DNA polymerase III sliding clamp | | 分子名称: | DNA polymerase III subunit beta | | 著者 | Gui, W.J, Lin, S.Q, Chen, Y.Y, Zhang, X.E, Bi, L.J, Jiang, T. | | 登録日 | 2010-09-30 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Crystal structure of DNA polymerase III beta sliding clamp from Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
7UA7
 
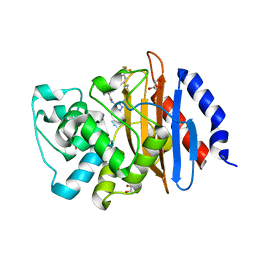 | | Crystal structure of indoline 9 with KPC-2 | | 分子名称: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, tert-butyl {(3R)-3-[(1H-tetrazol-5-yl)carbamoyl]-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indol-3-yl}carbamate | | 著者 | Akhtar, A, Chen, Y. | | 登録日 | 2022-03-11 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.247 Å) | | 主引用文献 | Probing the binding hot spots of KPC-2 carbapenemase using reversible tetrazole-based inhibitors
To Be Published
|
|
3QD7
 
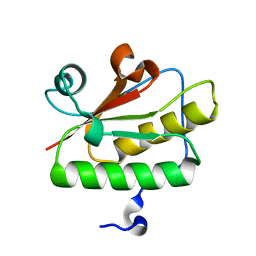 | | Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli | | 分子名称: | Uncharacterized protein ydaL | | 著者 | Gui, W.J, Qu, Q.H, Chen, Y.Y, Wang, M, Zhang, X.E, Bi, L.J, Jiang, T. | | 登録日 | 2011-01-18 | | 公開日 | 2011-06-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli.
J.Struct.Biol., 174, 2011
|
|
