1SDI
 
 | | 1.65 A structure of Escherichia coli ycfC gene product | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Hypothetical protein ycfC | | 著者 | Borek, D, Otwinowski, Z, Chen, Y, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-02-13 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural analysis of Escherichia coli ycfC gene product
To be Published
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | 分子名称: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | 登録日 | 2011-05-30 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | 分子名称: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | 著者 | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | 登録日 | 2011-05-30 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZT
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose | | 分子名称: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | 登録日 | 2011-05-30 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
4X3Q
 
 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in complex with SAH | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, SibL | | 著者 | liu, J.S, Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | 登録日 | 2014-12-01 | | 公開日 | 2015-11-25 | | 実験手法 | X-RAY DIFFRACTION (2.586 Å) | | 主引用文献 | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in complex with SAH
To Be Published, 2015
|
|
5IE1
 
 | | Crystal structure of BACE1 in complex with 3-(2-amino-6-(o-tolyl)quinolin-3-yl)-N-(3,3-dimethylbutyl)propanamide | | 分子名称: | 3-[2-amino-6-(2-methylphenyl)quinolin-3-yl]-N-(3,3-dimethylbutyl)propanamide, Beta-secretase 1, GLYCEROL, ... | | 著者 | Jordan, J.B, Whittington, D.A, Bartberger, M.D, Sickmier, E.A, Chen, K, Cheng, Y, Judd, T. | | 登録日 | 2016-02-24 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.298 Å) | | 主引用文献 | Fragment-Linking Approach Using (19)F NMR Spectroscopy To Obtain Highly Potent and Selective Inhibitors of beta-Secretase.
J.Med.Chem., 59, 2016
|
|
6LGW
 
 | | Structure of Rabies virus glycoprotein in complex with neutralizing antibody 523-11 at acidic pH | | 分子名称: | Glycoprotein, scFv 523-11 | | 著者 | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | 登録日 | 2019-12-06 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.9037 Å) | | 主引用文献 | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
6LGX
 
 | | Structure of Rabies virus glycoprotein at basic pH | | 分子名称: | Glycoprotein,Glycoprotein,Glycoprotein | | 著者 | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | 登録日 | 2019-12-06 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.097 Å) | | 主引用文献 | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
6LSA
 
 | | Complex structure of bovine herpesvirus 1 glycoprotein D and bovine nectin-1 IgV | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | 著者 | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | 登録日 | 2020-01-17 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.204 Å) | | 主引用文献 | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6LS9
 
 | | Crystal structure of bovine herpesvirus 1 glycoprotein D | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | 著者 | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | 登録日 | 2020-01-17 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.503 Å) | | 主引用文献 | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
1ZOI
 
 | | Crystal Structure of a Stereoselective Esterase from Pseudomonas putida IFO12996 | | 分子名称: | esterase | | 著者 | Elmi, F, Lee, H.T, Huang, J.Y, Hsieh, Y.C, Wang, Y.L, Chen, Y.J, Shaw, S.Y, Chen, C.J. | | 登録日 | 2005-05-13 | | 公開日 | 2006-05-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Stereoselective esterase from Pseudomonas putida IFO12996 reveals alpha/beta hydrolase folds for D-beta-acetylthioisobutyric acid synthesis
J.Bacteriol., 187, 2005
|
|
4JIS
 
 | | Crystal structure of ribitol 5-phosphate cytidylyltransferase (TarI) from Bacillus subtilis | | 分子名称: | ribitol-5-phosphate cytidylyltransferase | | 著者 | Yang, C.S, Chen, S.C, Chen, Y.R, Kuan, S.M, Liu, Y.H, Chen, Y. | | 登録日 | 2013-03-06 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.772 Å) | | 主引用文献 | Crystal structure of ribitol 5-phosphate cytidylyltransferase (TarI) from Bacillus subtilis
To be Published
|
|
1P2A
 
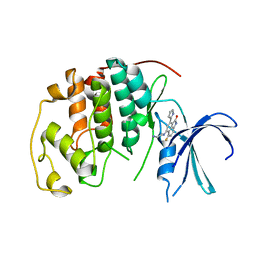 | | The structure of cyclin dependent kinase 2 (CKD2) with a trisubstituted naphthostyril inhibitor | | 分子名称: | 5-[(2-AMINOETHYL)AMINO]-6-FLUORO-3-(1H-PYRROL-2-YL)BENZO[CD]INDOL-2(1H)-ONE, Cell division protein kinase 2 | | 著者 | Liu, J.-J, Dermatakis, A, Lukacs, C.M, Konzelmann, F, Chen, Y, Kammlott, U, Depinto, W, Yang, H, Yin, X, Chen, Y, Schutt, A, Simcox, M.E, Luk, K.-C. | | 登録日 | 2003-04-15 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | 3,5,6-Trisubstituted Naphthostyrils as CDK2 Inhibitors
BIOORG.MED.CHEM., 13, 2003
|
|
1BC4
 
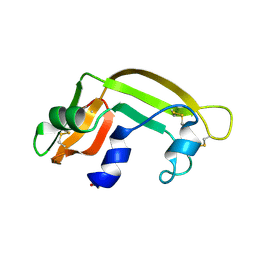 | | THE SOLUTION STRUCTURE OF A CYTOTOXIC RIBONUCLEASE FROM THE OOCYTES OF RANA CATESBEIANA (BULLFROG), NMR, 15 STRUCTURES | | 分子名称: | RIBONUCLEASE | | 著者 | Chang, C.-F, Chen, C, Chen, Y.-C, Hom, K, Huang, R.-F, Huang, T. | | 登録日 | 1998-05-05 | | 公開日 | 1998-10-14 | | 最終更新日 | 2019-12-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a cytotoxic ribonuclease from the oocytes of Rana catesbeiana (bullfrog).
J.Mol.Biol., 283, 1998
|
|
3CLH
 
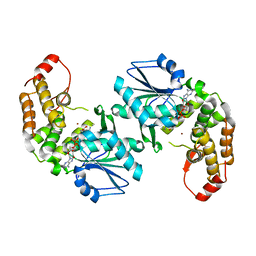 | | Crystal structure of 3-dehydroquinate synthase (DHQS)from Helicobacter pylori | | 分子名称: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Wang, W.C, Liu, J.S, Cheng, W.C, Wang, H.J, Chen, Y.C. | | 登録日 | 2008-03-19 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-based inhibitor discovery of Helicobacter pylori dehydroquinate synthase.
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2O14
 
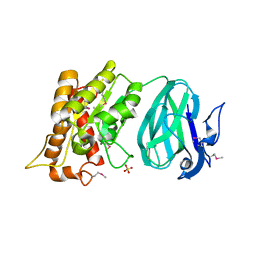 | | X-Ray Crystal Structure of Protein YXIM_BACsu from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR595 | | 分子名称: | Hypothetical protein yxiM, MANGANESE (II) ION, SULFATE ION | | 著者 | Kuzin, A.P, Chen, Y, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-11-28 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-Ray structure of the hypothetical protein YXIM_BACsu from Bacillus subtilis.
To be Published
|
|
3WRE
 
 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | 分子名称: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | 登録日 | 2014-02-25 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRG
 
 | | The complex structure of HypBA1 with L-arabinose | | 分子名称: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | 著者 | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | 登録日 | 2014-02-25 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
4RM0
 
 | | Crystal structure of Norovirus OIF P domain in complex with Lewis a trisaccharide | | 分子名称: | Capsid protein, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Liu, W, Chen, Y, Tan, M, Xia, M, Li, X, Jiang, X, Rao, Z. | | 登録日 | 2014-10-18 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface.
Plos Pathog., 11, 2015
|
|
8IVB
 
 | | K113-Ubiquitinated BAK | | 分子名称: | Bcl-2 homologous antagonist/killer, Ubiquitin | | 著者 | Dong, X, Cheng, P, Hou, Y.Z, Chen, Y.K, Liu, Z. | | 登録日 | 2023-03-26 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Parkin-mediated ubiquitination inhibits BAK apoptotic activity by blocking its canonical hydrophobic groove.
Commun Biol, 6, 2023
|
|
4RLZ
 
 | | Crystal structure of Norovirus OIF P domain | | 分子名称: | Capsid protein, GLYCEROL | | 著者 | Liu, W, Chen, Y, Tan, M, Xia, M, Li, X, Jiang, X, Rao, Z. | | 登録日 | 2014-10-18 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface.
Plos Pathog., 11, 2015
|
|
6XFN
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW243 | | 分子名称: | 3C-like proteinase, GLYCEROL, UAW243 | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-06-15 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW247 | | 分子名称: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-06-06 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | 分子名称: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
4UAI
 
 | |
