8K12
 
 | | SID1 transmembrane family member 2 | | Descriptor: | SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K1B
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K10
 
 | | SID1 transmembrane family member 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K13
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K1D
 
 | | SID1 transmembrane family member 1 | | Descriptor: | SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8IAZ
 
 | | Cryo-EM structure of the ISFba1 TnpB-reRNA-dsDNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*AP*TP*GP*GP*AP*CP*CP*AP*TP*CP*AP*GP*CP*TP*CP*CP*TP*AP*AP*TP*GP*G)-3'), DNA (5'-D(P*CP*CP*AP*TP*TP*AP*GP*GP*AP*GP*CP*TP*GP*AP*TP*G)-3'), RNA (207-MER), ... | | Authors: | Yin, M, Zhou, F, Zhu, Y, Huang, Z. | | Deposit date: | 2023-02-09 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Discovery and structural mechanism of DNA endonucleases guided by RAGATH-18-derived RNAs.
Cell Res., 34, 2024
|
|
2WW1
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with Thiomannobioside | | Descriptor: | PUTATIVE ALPHA-1,2-MANNOSIDASE, alpha-D-mannopyranose-(1-2)-methyl 2-thio-alpha-D-mannopyranoside | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
4I11
 
 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates. | | Descriptor: | Beta-secretase 1, N-(3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)-L-phenylalanine, ZINC ION | | Authors: | Bowers, B, Xu, Y, Yuan, S, Probst, G.D, Hom, R.K, Chan, W, Konradi, A.W, Sham, H.L, Zhu, Y.L, Beroza, P, Pan, H, Brecht, E, Yao, N, Lougheed, J, Artis, D.R, Tam, D, Bova, M. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7ED5
 
 | | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Shannon, A, Fattorini, V, Sama, B, Selisko, B, Feracci, M, Falcou, C, Gauffre, P, El Kazzi, P, Delpal, A, Decroly, E, Alvarez, K, Eydoux, C, Guillemot, J.-C, Moussa, A, Good, S, Colla, P, Lin, K, Sommadossi, J.-P, Zhu, Y.X, Yan, X.D, Shi, H, Ferron, F, Canard, B. | | Deposit date: | 2021-03-15 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase.
Nat Commun, 13, 2022
|
|
4FMA
 
 | | EspG structure | | Descriptor: | ACETIC ACID, EspG protein, FORMIC ACID, ... | | Authors: | Shao, F, Zhu, Y, Hu, L. | | Deposit date: | 2012-06-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structurally Distinct Bacterial TBC-like GAPs Link Arf GTPase to Rab1 Inactivation to Counteract Host Defenses.
Cell(Cambridge,Mass.), 150, 2012
|
|
7W2O
 
 | |
2WW3
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with thiomannobioside | | Descriptor: | CALCIUM ION, GLYCEROL, PUTATIVE ALPHA-1,2-MANNOSIDASE, ... | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic Insights Into a Ca(2+)-Dependent Family of Alpha-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
2WVX
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | CALCIUM ION, GLYCEROL, PUTATIVE ALPHA-1,2-MANNOSIDASE | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
6JC3
 
 | | The Cryo-EM structure of nucleoprotein-RNA complex of Newcastle disease virus | | Descriptor: | Nucleocapsid, polyU | | Authors: | Song, X, Shan, H, Zhu, Y, Ding, W, Ouyang, S, Shen, Q.T, Liu, Z.J. | | Deposit date: | 2019-01-28 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Self-capping of nucleoprotein filaments protects the Newcastle disease virus genome.
Elife, 8, 2019
|
|
4EAY
 
 | | Crystal structures of mannonate dehydratase from Escherichia coli strain K12 complexed with D-mannonate | | Descriptor: | CHLORIDE ION, D-MANNONIC ACID, MANGANESE (II) ION, ... | | Authors: | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
5XLX
 
 | |
5XN7
 
 | |
4EF5
 
 | | Crystal structure of STING CTD | | Descriptor: | Transmembrane protein 173 | | Authors: | Ouyang, S, Ru, H, Shaw, N, Jiang, Y, Niu, F, Zhu, Y, Qiu, W, Li, Y, Liu, Z.-J. | | Deposit date: | 2012-03-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding
Immunity, 36, 2012
|
|
5Y8E
 
 | |
6JAU
 
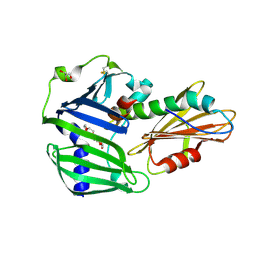 | | The complex structure of Pseudomonas aeruginosa MucA/MucB. | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Li, T, He, L.H, Li, C.C, Liu, L, Peng, C.T, Shen, Y.L, Qin, X.F, Xiao, Q.J, Zhu, Y.B, Song, Y.J, Zhao, N.l, Zhao, C, Yang, J, Mu, X.Y, Huang, Q, Bao, R. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Molecular basis of the lipid-induced MucA-MucB dissociation in Pseudomonas aeruginosa.
Commun Biol, 3, 2020
|
|
5Y8F
 
 | |
6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | Descriptor: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | Deposit date: | 2019-04-24 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
6A0A
 
 | |
5XLY
 
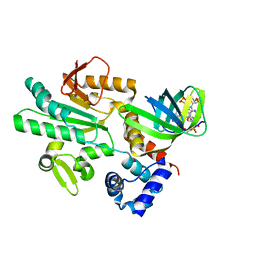 | | Crystal structure of CheR1 in complex with c-di-GMP-bound MapZ | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Chemotaxis protein methyltransferase 1, Cyclic diguanosine monophosphate-binding protein PA4608 | | Authors: | Yuan, Z, Zhu, Y, Gu, L. | | Deposit date: | 2017-05-12 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Structural basis for the regulation of chemotaxis by MapZ in the presence of c-di-GMP
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6A0C
 
 | | Structure of a triple-helix region of human collagen type III | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, collagen type III peptide | | Authors: | Yang, X, Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Characterization by high-resolution crystal structure analysis of a triple-helix region of human collagen type III with potent cell adhesion activity.
Biochem. Biophys. Res. Commun., 508, 2019
|
|
