7JMW
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain in complex with cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
6PZY
 
 | | CryoEM derived model of NA-73 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-73 fragment antibody heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6X1Q
 
 | | 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, SODIUM ION | | Authors: | Merk, A, Fukumura, T, Zhu, X, Darling, J, Grisshammer, R, Ognjenovic, J, Subramaniam, S. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | 1.8 angstrom resolution structure of beta-galactosidase with a 200 kV CRYO ARM electron microscope.
Iucrj, 7, 2020
|
|
6Y6I
 
 | | lipopolysaccharide outer core galactosyltransferase WaaB and UDP complex | | Descriptor: | Lipopolysaccharide 1,6-galactosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Z.Y, Ashworth, G, Wang, Z.S, Zhu, X.F, Dong, C.J. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into the lipopolysaccharide outer core galactosyltransferase WaaB
To Be Published
|
|
6Y6G
 
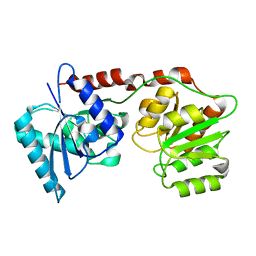 | | lipopolysaccharide outer core galactosyltransferase WaaB apo form | | Descriptor: | Lipopolysaccharide 1,6-galactosyltransferase | | Authors: | Zhang, Z.Y, Ashworth, G, Wang, Z.S, Zhu, X.F, Dong, C.J. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the lipopolysaccharide outer core galactosyltransferase WaaB
To Be Published
|
|
6XC4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 heavy chain, CC12.3 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XC2
 
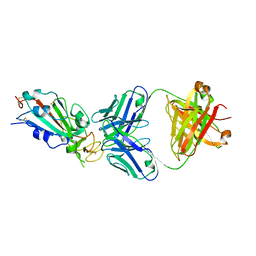 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
8SEU
 
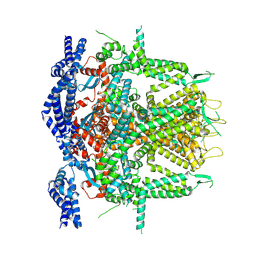 | | Cryo-EM Structure of RyR1 (Local Refinement of TMD) | | Descriptor: | Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
4BS9
 
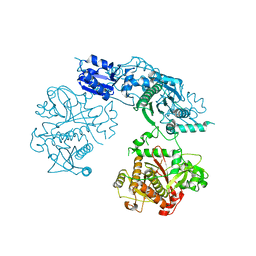 | | Structure of the heterocyclase TruD | | Descriptor: | TRUD, ZINC ION | | Authors: | Koehnke, J, Bent, A.F, Zollman, D, Smith, K, Houssen, W.E, Zhu, X, Mann, G, Lebl, T, Scharff, R, Shirran, S, Botting, C.H, Jaspars, M, Schwarz-Linek, U, Naismith, J.H. | | Deposit date: | 2013-06-09 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Cyanobactin Heterocyclase Enzyme: A Processive Adenylase that Operates with a Defined Order of Reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6XKP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-270 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-270 Heavy Chain, CV07-270 Light Chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
8SEV
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S (Local Refinement of TMD) | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEY
 
 | | Cryo-EM Structure of RyR1 + Adenosine (Local Refinement of TMD) | | Descriptor: | ADENOSINE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEW
 
 | | Cryo-EM Structure of RyR1 + ADP (Local Refinement of TMD) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SF0
 
 | | Cryo-EM Structure of RyR1 + cAMP (Local Refinement of TMD) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEZ
 
 | | Cryo-EM Structure of RyR1 + Adenine (Local Refinement of TMD) | | Descriptor: | ADENINE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEX
 
 | | Cryo-EM Structure of RyR1 + AMP (Local Refinement of TMD) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
6XKQ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-250 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-250 Heavy Chain, CV07-250 Light Chain, ... | | Authors: | Yuan, M, Liu, H, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
7TP4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K398.22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, K398.22 heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7LM9
 
 | | Crystal structure of SARS-CoV spike protein receptor-binding domain in complex with a cross-neutralizing antibody CV38-142 Fab isolated from COVID-19 patient | | Descriptor: | 1,2-ETHANEDIOL, CV38-142 Fab heavy chain, CV38-142 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A combination of cross-neutralizing antibodies synergizes to prevent SARS-CoV-2 and SARS-CoV pseudovirus infection.
Cell Host Microbe, 29, 2021
|
|
3ME2
 
 | | Crystal structure of mouse RANKL-RANK complex | | Descriptor: | CHLORIDE ION, SODIUM ION, Tumor necrosis factor ligand superfamily member 11, ... | | Authors: | Walter, S.W, Liu, C.Z, Zhu, X.K, Wu, Y, Owens, R.J, Stuart, D.I, Gao, B, Ren, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Insights of RANKL-RANK Interaction and Signaling.
J.Immunol., 2010
|
|
9CUO
 
 | | Crystal structure of CRBN with compound 3 | | Descriptor: | (3S)-3-(3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, Protein cereblon, ... | | Authors: | Zheng, X, Ji, N, Campbell, V, Slavin, A, Zhu, X, Chen, D, Rong, H, Enerson, B, Mayo, M, Sharma, K, Browne, C.M, Klaus, C.R, Li, H, Massa, G, McDonald, A.A, Shi, Y, Sintchak, M, Skouras, S, Walther, D.M, Yuan, K, Zhang, Y, Kelleher, J, Guang, L, Luo, X, Mainolfi, N, Weiss, M.M. | | Deposit date: | 2024-07-26 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of KT-474─a Potent, Selective, and Orally Bioavailable IRAK4 Degrader for the Treatment of Autoimmune Diseases.
J.Med.Chem., 2024
|
|
3ME4
 
 | | Crystal structure of mouse RANK | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walter, S.W, Liu, C, Zhu, X, Wu, Y, Owens, R.J, Stuart, D.I, Gao, B, Ren, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Functional Insights of RANKL-RANK Interaction and Signaling.
J.Immunol., 2010
|
|
3LZD
 
 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | Descriptor: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
7JMX
 
 | | Crystal structure of a SARS-CoV-2 cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | ACETATE ION, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
5EQY
 
 | | Crystal structure of choline kinase alpha-1 bound by 5-[(4-methyl-1,4-diazepan-1-yl)methyl]-2-[4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]benzenecarbonitrile (compound 65) | | Descriptor: | 5-[(4-methyl-1,4-diazepan-1-yl)methyl]-2-[4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]benzenecarbonitrile, Choline kinase alpha | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
