3SND
 
 | |
3SNC
 
 | |
3SNE
 
 | |
3SN8
 
 | |
3SNB
 
 | |
3SNA
 
 | |
4JTT
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 066 | | Descriptor: | 2,4-dimethyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Zhu, J, Ren, X. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
TO BE PUBLISHED
|
|
4JTS
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 072 | | Descriptor: | 4-methyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
To be Published
|
|
4JTU
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with brequinar analogue | | Descriptor: | 6-bromo-2-{4-[(2R)-butan-2-yl]phenyl}-3-methylquinoline-4-carboxylic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
To be Published
|
|
4JGD
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A016 | | Descriptor: | 1-[4-methyl-2-(naphthalen-2-ylamino)-1,3-thiazol-5-yl]ethanone, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-01 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A016
TO BE PUBLISHED
|
|
4JS3
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057 | | Descriptor: | 2-chloro-4-methyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETIC ACID, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Zhu, J, Ren, X. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
TO BE PUBLISHED
|
|
7KXT
 
 | | Crystal structure of human EED | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
5GKA
 
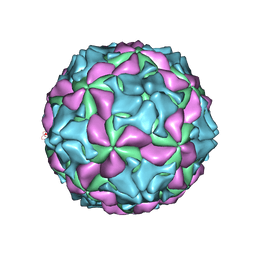 | | cryo-EM structure of human Aichi virus | | Descriptor: | Genome polyprotein, capsid protein VP0, capsid protein VP1 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Tuthill, T.J, Fry, E.E, Rao, Z.H, Stuart, D.I. | | Deposit date: | 2016-07-04 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of human Aichi virus and implications for receptor binding
Nat Microbiol, 1, 2016
|
|
6M2B
 
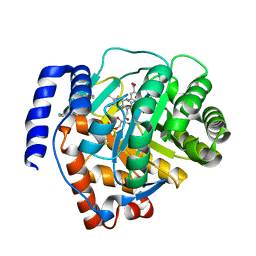 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with S416 | | Descriptor: | 2-[(E)-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]-methyl-hydrazinylidene]methyl]benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Li, H. | | Deposit date: | 2020-02-27 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2.
Protein Cell, 11, 2020
|
|
3TIU
 
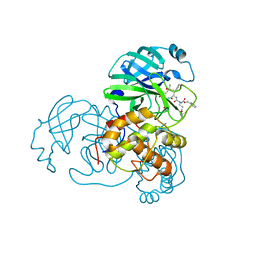 | | Crystal structure of SARS coronavirus main protease complexed with an alpha,beta-unsaturated ethyl ester inhibitor SG82 | | Descriptor: | DIMETHYL SULFOXIDE, ETHYL (5S,8S,11R)-8-BENZYL-5-(3-TERT-BUTOXY-3-OXOPROPYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE, SARS coronavirus main protease | | Authors: | Zhu, L, Hilgenfeld, R. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of SARS-Cov main protease complexed with a series of peptidic unsaturated esters
To be Published
|
|
3TIT
 
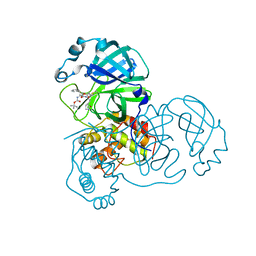 | |
3TNT
 
 | |
3TNS
 
 | | Crystal structure of SARS coronavirus main protease complexed with an alpha, beta-unsaturated ethyl ester inhibitor SG83 | | Descriptor: | ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE, SARS coronavirus main protease | | Authors: | Zhu, L, Hilgenfeld, R. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of SARS-Cov main protease complexed with a series of peptidic unsaturated esters
To be Published
|
|
6M6E
 
 | |
6M6F
 
 | |
6BA6
 
 | | Solution structure of Rap1b/talin complex | | Descriptor: | Ras-related protein Rap-1b, Talin-1 | | Authors: | Zhu, L, Yang, J, Qin, J. | | Deposit date: | 2017-10-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Rap1b bound to talin reveals a pathway for triggering integrin activation.
Nat Commun, 8, 2017
|
|
8A2G
 
 | | Crystal structure of Sebokelevirus 2A2 protein | | Descriptor: | 1,2-ETHANEDIOL, 2A2 protein, TETRAETHYLENE GLYCOL | | Authors: | Zhu, L, Von Castelmur, E, Whang, X, Ren, J, Fry, E, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family
to be published
|
|
5ZID
 
 | | Crystal Structure of human DPP-IV in complex with HL2 | | Descriptor: | (2S,3R)-2-amino-9-methoxy-3-(2,4,5-trifluorophenyl)-2,3-dihydro-1H-naphtho[2,1-b]pyran-8-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Zhu, L, Li, H, Wu, F. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of human DPP-IV in complex with HL2
To Be Published
|
|
8JPX
 
 | | Cryo-EM structure of PfAgo-guide DNA-target DNA complex | | Descriptor: | Excess DNA, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
8WD8
 
 | | Cryo-EM structure of TtdAgo-guide DNA-target DNA complex | | Descriptor: | Argonaute family protein, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
