1YIS
 
 | | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase | | Descriptor: | SULFATE ION, adenylosuccinate lyase | | Authors: | Symersky, J, Schormann, N, Lu, S, Zhang, Y, Karpova, E, Qiu, S, Huang, W, Cao, Z, Zhou, J, Luo, M, Arabshahi, A, McKinstry, A, Luan, C.-H, Luo, D, Johnson, D, An, J, Tsao, J, Delucas, L, Shang, Q, Gray, R, Li, S, Bray, T, Chen, Y.-J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase
To be Published
|
|
8JNY
 
 | |
8JNZ
 
 | | Human ADP-ribosyltransferase 1 (PARP1) catalytic domain bound to a pyrazolopyrimidine carboxamide inhibitor | | Descriptor: | 6-methylpyrazolo[1,5-a]pyrimidine-3-carboxamide, Poly [ADP-ribose] polymerase 1, processed C-terminus, ... | | Authors: | Wang, X.Y, Wang, C.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Human ADP-ribosyltransferase 1 (PARP1) catalytic domain bound to a pyrazolopyrimidine carboxamide inhibitor
To Be Published
|
|
3CM8
 
 | | A RNA polymerase subunit structure from virus | | Descriptor: | Polymerase acidic protein, peptide from RNA-directed RNA polymerase catalytic subunit | | Authors: | He, X, Zhou, J, Zeng, Z, Ma, J, Zhang, R, Rao, Z, Liu, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Crystal structure of the polymerase PAC-PB1N complex from an avian influenza H5N1 virus
Nature, 454, 2008
|
|
3DON
 
 | | Crystal structure of shikimate dehydrogenase from Staphylococcus epidermidis | | Descriptor: | GLYCEROL, Shikimate dehydrogenase | | Authors: | Han, C, Hu, T, Wu, D, Zhou, J, Shen, X, Qu, D, Jiang, H. | | Deposit date: | 2008-07-05 | | Release date: | 2009-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Febs J., 276, 2009
|
|
3DOO
 
 | | Crystal structure of shikimate dehydrogenase from Staphylococcus epidermidis complexed with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | Authors: | Han, C, Hu, T, Wu, D, Zhou, J, Shen, X, Qu, D, Jiang, H. | | Deposit date: | 2008-07-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Febs J., 276, 2009
|
|
6LY4
 
 | | The crystal structure of the BM3 mutant LG-23 in complex with testosterone | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, ... | | Authors: | Peng, Y, Chen, J, Zhou, J, Li, A, ReetZ, M.T. | | Deposit date: | 2020-02-13 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Regio- and Stereoselective Steroid Hydroxylation at C7 by Cytochrome P450 Monooxygenase Mutants.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3OPW
 
 | | Crystal Structure of the Rph1 catalytic core | | Descriptor: | DNA damage-responsive transcriptional repressor RPH1 | | Authors: | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
3OPT
 
 | | Crystal structure of the Rph1 catalytic core with a-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DNA damage-responsive transcriptional repressor RPH1, NICKEL (II) ION | | Authors: | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
5KPP
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[(3R)-3-methyl-4-[2,2,2-tris(fluoranyl)ethyl]piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5KPO
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[3-(4-ethyl-3-oxidanylidene-piperazin-1-yl)carbonyl-4-fluoranyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Yao, H.P, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5KPQ
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[(3R)-3-methyl-4-propyl-piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5KPN
 
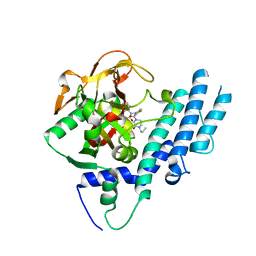 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-propyl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Yao, H.P, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | Descriptor: | D-Cysteine desulfhydrase | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
3MGT
 
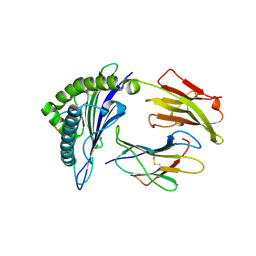 | | Crystal structure of a H5-specific CTL epitope variant derived from H5N1 influenza virus in complex with HLA-A*0201 | | Descriptor: | 10-meric peptide from Hemagglutinin, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Sun, Y, Liu, J, Yang, M, Gao, F, Zhou, J, Kitamura, Y. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses
J.Gen.Virol., 91, 2010
|
|
3H5V
 
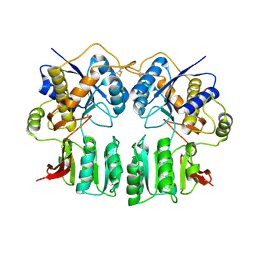 | | Crystal structure of the GluR2-ATD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
1ZCY
 
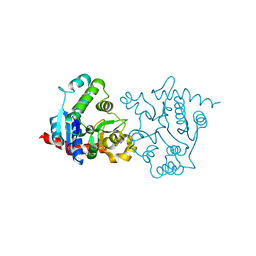 | | apo form of a mutant of glycogenin in which Asp159 is replaced by Ser | | Descriptor: | Glycogenin-1, SULFATE ION | | Authors: | Hurley, T.D, Stout, S.L, Miner, E, Zhou, J, Roach, P.J. | | Deposit date: | 2005-04-13 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Requirements for catalysis in mammalian glycogenin.
J.Biol.Chem., 280, 2005
|
|
1ZCU
 
 | | apo form of the 162S mutant of glycogenin | | Descriptor: | Glycogenin-1 | | Authors: | Hurley, T.D, Stout, S.L, Miner, E, Zhou, J, Roach, P.J. | | Deposit date: | 2005-04-13 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Requirements for catalysis in mammalian glycogenin.
J.Biol.Chem., 280, 2005
|
|
1YGH
 
 | | HAT DOMAIN OF GCN5 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | GLYCEROL, PROTEIN (TRANSCRIPTIONAL ACTIVATOR GCN5) | | Authors: | Trievel, R.C, Rojas, J.R, Sterner, D.E, Venkataramani, R, Wang, L, Zhou, J, Allis, C.D, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-27 | | Release date: | 1999-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3MGO
 
 | | Crystal structure of a H5-specific CTL epitope derived from H5N1 influenza virus in complex with HLA-A*0201 | | Descriptor: | 10-meric peptide from Hemagglutinin, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Sun, Y, Liu, J, Yang, M, Gao, F, Zhou, J, Kitamura, Y. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses
J.Gen.Virol., 91, 2010
|
|
1ZCT
 
 | | structure of glycogenin truncated at residue 270 in a complex with UDP | | Descriptor: | Glycogenin-1, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Hurley, T.D, Stout, S.L, Miner, E, Zhou, J, Roach, P.J. | | Deposit date: | 2005-04-13 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Requirements for catalysis in mammalian glycogenin.
J.Biol.Chem., 280, 2005
|
|
1ZDF
 
 | | Ser162 mutant of glycogenin complexed with UDP-glucose and manganese | | Descriptor: | Glycogenin-1, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hurley, T.D, Stout, S.L, Miner, E, Zhou, J, Roach, P.J. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Requirements for catalysis in mammalian glycogenin.
J.Biol.Chem., 280, 2005
|
|
3H5W
 
 | | Crystal structure of the GluR2-ATD in space group P212121 without solvent | | Descriptor: | Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
1ZCV
 
 | | apo form of a mutant of glycogenin in which Asp159 is replaced by Asn | | Descriptor: | Glycogenin-1, SULFATE ION | | Authors: | Hurley, T.D, Stout, S.L, Miner, E, Zhou, J, Roach, P.J. | | Deposit date: | 2005-04-13 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Requirements for catalysis in mammalian glycogenin.
J.Biol.Chem., 280, 2005
|
|
