6KNI
 
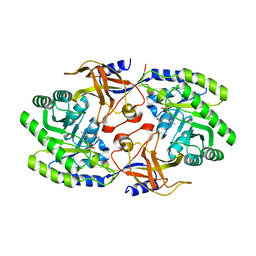 | |
5X4J
 
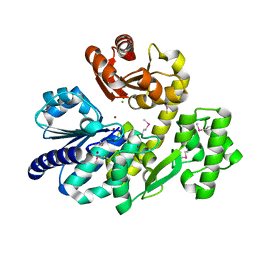 | | The crystal structure of Pyrococcus furiosus RecJ (Zn-soaking) | | Descriptor: | CHLORIDE ION, Uncharacterized protein, ZINC ION | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4K
 
 | | The complex crystal structure of Pyrococcus furiosus RecJ and CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Uncharacterized protein, ZINC ION | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4H
 
 | | The crystal structure of Pyrococcus furiosus RecJ (wild-type) | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
6K7Y
 
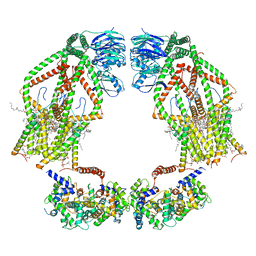 | | Intact human mitochondrial calcium uniporter complex with MICU1/MICU2 subunits | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhuo, W, Zhou, H, Yang, M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
5X4I
 
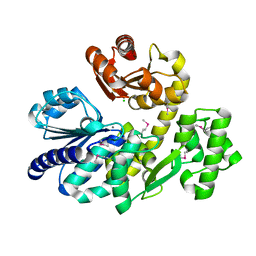 | | Pyrococcus furiosus RecJ (D83A, Mn-soaking) | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | Descriptor: | ZIKV NS1 | | Authors: | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
7VRC
 
 | |
5ZBA
 
 | | Crystal structure of Rtt109-Asf1-H3-H4-CoA complex | | Descriptor: | COENZYME A, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
5Z9E
 
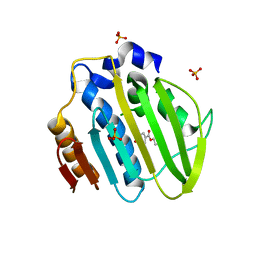 | |
5Z9M
 
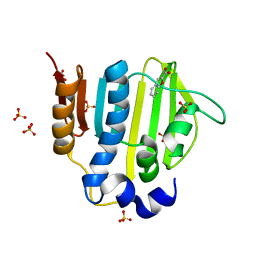 | |
5Z9B
 
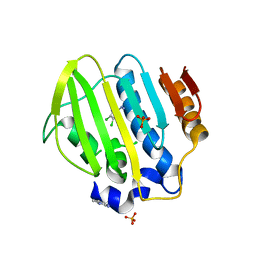 | | Bacterial GyrB ATPase domain in complex with (3,4-dichlorophenyl)hydrazine | | Descriptor: | (3,4-dichlorophenyl)hydrazine, 1H-benzimidazol-2-amine, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-02 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z4H
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 4-(4-hydroxyphenyl)sulfanylphenol, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-01-11 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5ZBB
 
 | | Crystal structure of Rtt109-Asf1-H3-H4 complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
5Z9P
 
 | |
5Z9F
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 2-hydroxybenzonitrile, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9L
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 2-fluoro-4-hydroxybenzonitrile, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9Q
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 5-phenyl-1H-pyrazol-3-amine, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z4O
 
 | |
5XSG
 
 | | Ultrahigh resolution structure of FUS (37-42) SYSGYS determined by MicroED | | Descriptor: | RNA-binding protein FUS | | Authors: | Luo, F, Gui, X, Zhou, H, Li, D, Li, X, Liu, C. | | Deposit date: | 2017-06-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.73 Å) | | Cite: | Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6J60
 
 | | hnRNP A1 reversible amyloid core GFGGNDNFG (residues 209-217) | | Descriptor: | 9-mer peptide (GFGGNDNFG) from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Luo, F, Zhou, H, Gui, X, Li, D, Li, X, Liu, C. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.96 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5Z9N
 
 | |
5Z10
 
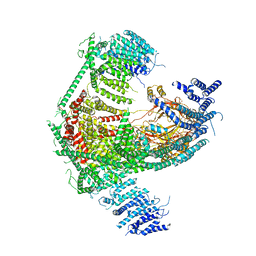 | | Structure of the mechanosensitive Piezo1 channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhao, Q, Zhou, H, Chi, S, Wang, Y, Wang, J, Geng, J, Wu, K, Liu, W, Zhang, T, Dong, M.-Q, Wang, J, Li, X, Xiao, B. | | Deposit date: | 2017-12-22 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structure and mechanogating mechanism of the Piezo1 channel.
Nature, 554, 2018
|
|
5ZAN
 
 | |
5ZNK
 
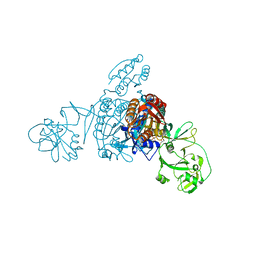 | | Crystal structure of a bacterial ProRS with ligands | | Descriptor: | 7-chloro-6-fluoro-3-{2-oxo-3-[(2S)-piperidin-2-yl]propyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cheng, B, Yu, Y, Zhou, H. | | Deposit date: | 2018-04-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
