4XA1
 
 | | Crystal Structure of the coiled-coil surrounding Skip 1 of MYH7 | | Descriptor: | Gp7-MYH7(1173-1238)-EB1 chimera protein | | Authors: | Taylor, K.C, Buvoli, M, Korkmaz, E.N, Buvoli, A, Zheng, Y, Heinz, N.T, Qiang, C, Leinwand, L.A, Rayment, I. | | Deposit date: | 2014-12-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5DBI
 
 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NAD+ and 10-oxogeranial | | Descriptor: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, Iridoid synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hu, Y.M, Liu, W.D, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | Descriptor: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | Authors: | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
5DBG
 
 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NAD+ | | Descriptor: | Iridoid synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, W.D, Hu, Y.M, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8I83
 
 | |
8IKU
 
 | |
7DVJ
 
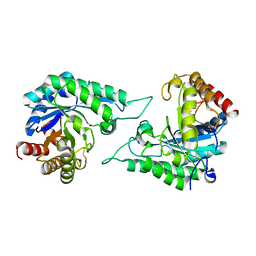 | | Structure of beta-mannanase BaMan113A with mannobiose | | Descriptor: | Endo-beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Liu, W.T, Liu, W.D, Zheng, Y.Y. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and structural investigation of a novel beta-mannanase BaMan113A from Bacillus sp. N16-5.
Int.J.Biol.Macromol., 182, 2021
|
|
7DV7
 
 | |
5CUU
 
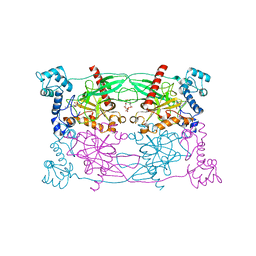 | | Crystal structure of Trypanosoma cruzi Vacuolar Soluble Pyrophosphatases in complex with bisphosphonate inhibitor BPH-1260 | | Descriptor: | 1-butyl-3-(2-hydroxy-2,2-diphosphonoethyl)-1H-imidazol-3-ium, Acidocalcisomal pyrophosphatase, D-MALATE, ... | | Authors: | Liu, W.D, Yang, Y.Y, Ko, T.P, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-07-25 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of Trypanosoma cruzi protein in complex with ligand
Acs Chem.Biol., 2016
|
|
8J0I
 
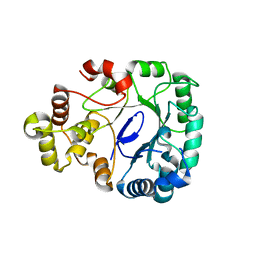 | | Aldo-keto reductase KmAKR | | Descriptor: | NADPH-dependent alpha-keto amide reductase, SODIUM ION | | Authors: | Xu, S.Y, Zhou, L, Xu, Y, Wang, Y.J, Zheng, Y.G. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Aldo-keto reductase KmAKR from Kluyveromyces marxianus
To Be Published
|
|
5CUY
 
 | | Crystal structure of Trypanosoma brucei Vacuolar Soluble Pyrophosphatases in apo form | | Descriptor: | Acidocalcisomal pyrophosphatase, CITRIC ACID, MAGNESIUM ION | | Authors: | Yang, Y.Y, Ko, T.P, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-07-25 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi protein in complex with ligand
Acs Chem.Biol., 2016
|
|
5DBF
 
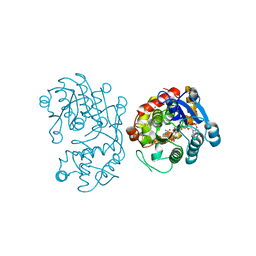 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NADPH | | Descriptor: | Iridoid synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hu, Y.M, Liu, W.D, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7ESC
 
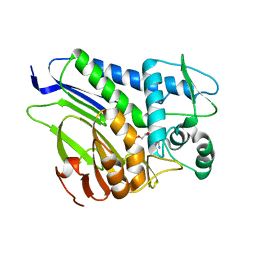 | | FmnB complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-05-10 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7F39
 
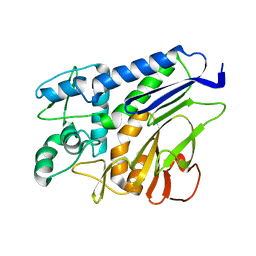 | | The structure of flavin transferase FmnB | | Descriptor: | FAD:protein FMN transferase | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-06-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
6B7N
 
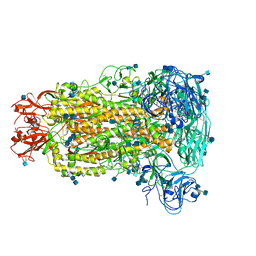 | | Cryo-electron microscopy structure of porcine delta coronavirus spike protein in the pre-fusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Tai, W, Du, L, Zhou, Y, Zhang, W, Li, F. | | Deposit date: | 2017-10-04 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy Structure of Porcine Deltacoronavirus Spike Protein in the Prefusion State
J. Virol., 92, 2018
|
|
7FB8
 
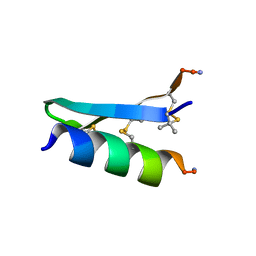 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd1 | | Descriptor: | ASP-ASP-LYS-ASP-CYS-ASP-GLU-TYR-CYS-LYS-LYS-THR-LYS-GLU-NH2, GLU-LE1-THR-GLY-HIS-ILE-GLU-GLY-PRO-THR-LE1-THR-LE1-HIS-CYS-LYS-NH2 | | Authors: | Yao, H, Yao, S, Zheng, Y, Moyer, A, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
7FBA
 
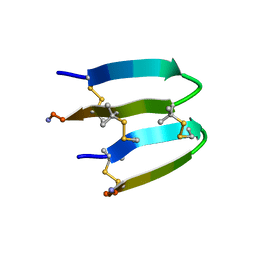 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd2 | | Descriptor: | ALA-LE1-CYS-GLU-CYS-GLY-PRO-THR-ARG-GLU-CYS-LYS-NH2, GLU-CYS-ARG-GLU-TYR-GLY-PRO-LE1-LYS-LE1-LE1-ALA-NH2 | | Authors: | Yao, H, Yao, S, Moyer, A, Zheng, Y, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
4LX3
 
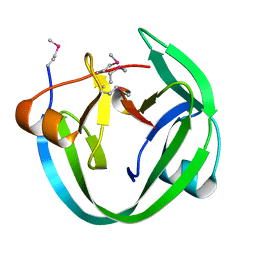 | | Conserved Residues that Modulate Protein trans-Splicing of Npu DnaE Split Intein | | Descriptor: | DNA polymerase III, alpha subunit, Nucleic acid binding, ... | | Authors: | Wu, Q, Gao, Z, Wei, Y, Ma, G, Zheng, Y, Dong, Y, Liu, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved residues that modulate protein trans-splicing of Npu DnaE split intein.
Biochem.J., 461, 2014
|
|
5CUX
 
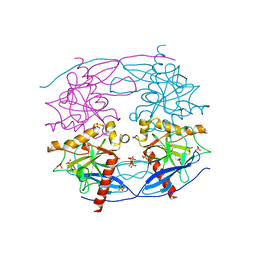 | | Crystal structure of N-terminal domain truncated Trypanosoma cruzi Vacuolar Soluble Pyrophosphatases in complex with PPi | | Descriptor: | Acidocalcisomal pyrophosphatase, PHOSPHATE ION, PYROPHOSPHATE 2- | | Authors: | Liu, W.D, Yang, Y.Y, Ko, T.P, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-07-25 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Trypanosoma cruzi protein in complex with ligand
Acs Chem.Biol., 2016
|
|
7OUO
 
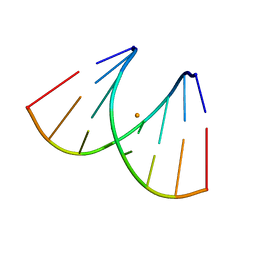 | | Crystal structure of RNA duplex [UCGUGCGA]2 in complex with Ba2+ cation | | Descriptor: | BARIUM ION, RNA (5'-R(*UP*CP*GP*UP*GP*CP*GP*A)-3') | | Authors: | Ruszkowski, M, Mao, S, Zheng, Y.Y, Ruszkowska, A, Sheng, J. | | Deposit date: | 2021-06-12 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Structural Insights Into the 5'UG/3'GU Wobble Tandem in Complex With Ba 2+ Cation.
Front Mol Biosci, 8, 2021
|
|
5MLE
 
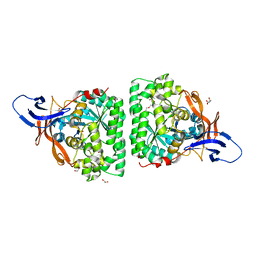 | | Crystal Structure of Human Dihydropyrimidinease-like 2 (DPYSL2A)/Collapsin Response Mediator Protein (CRMP2 13-516) Mutant Y479E/Y499E | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dihydropyrimidinase-related protein 2, ... | | Authors: | Sethi, R, Zheng, Y, Talon, R, Velupillai, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ahmed, A.A, von Delft, F. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Tuning microtubule dynamics to enhance cancer therapy by modulating FER-mediated CRMP2 phosphorylation.
Nat Commun, 9, 2018
|
|
7F2U
 
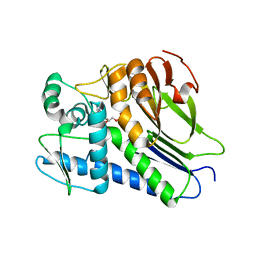 | | FmnB complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-06-14 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7C2J
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (without additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
5H9D
 
 | | Crystal structure of Heptaprenyl Diphosphate Synthase from Staphylococcus aureus | | Descriptor: | C-terminal peptide from Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, Farnesyl pyrophosphate synthetase, Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, ... | | Authors: | Wei, H.L, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure, Function, and Inhibition of Staphylococcus aureus Heptaprenyl Diphosphate Synthase
ChemMedChem, 11, 2016
|
|
