1KMI
 
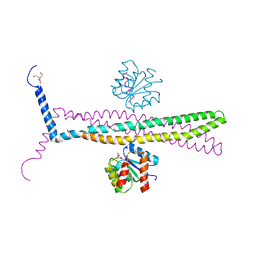 | | CRYSTAL STRUCTURE OF AN E.COLI CHEMOTAXIS PROTEIN, CHEZ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BICINE, Chemotaxis protein cheY, ... | | Authors: | Zhao, R, Collins, E.J, Bourret, R.B, Silversmith, R.E. | | Deposit date: | 2001-12-16 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ.
Nat.Struct.Biol., 9, 2002
|
|
4EGC
 
 | |
1RHI
 
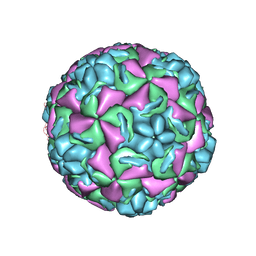 | | HUMAN RHINOVIRUS 3 COAT PROTEIN | | Descriptor: | CALCIUM ION, HUMAN RHINOVIRUS 3 COAT PROTEIN | | Authors: | Zhao, R, Rossmann, M.G. | | Deposit date: | 1996-06-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human rhinovirus 3 at 3.0 A resolution.
Structure, 4, 1996
|
|
8J5J
 
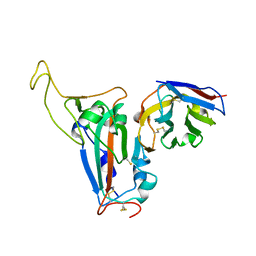 | | The crystal structure of bat coronavirus RsYN04 RBD bound to the antibody S43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nano antibody S43 | | Authors: | Zhao, R.C, Niu, S, Han, P, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2023-04-23 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.
Zool.Res., 44, 2023
|
|
8JIX
 
 | |
1T5I
 
 | | Crystal structure of the C-terminal domain of UAP56 | | Descriptor: | C_TERMINAL DOMAIN OF A PROBABLE ATP-DEPENDENT RNA HELICASE | | Authors: | Zhao, R, Green, M.R, Shen, J. | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of UAP56, a "DEXD/H-box" protein involved in pre-mRNA splicing and mRNA export
Structure, 12, 2004
|
|
1BZ9
 
 | |
1T6N
 
 | | Crystal structure of the N-terminal domain of human UAP56 | | Descriptor: | CITRATE ANION, Probable ATP-dependent RNA helicase | | Authors: | Zhao, R. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of UAP56, a DExD/H-Box Protein Involved in Pre-mRNA Splicing and mRNA Export
Structure, 12, 2004
|
|
6IYA
 
 | |
1B0G
 
 | |
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5WWP
 
 | | Crystal structure of Middle East respiratory syndrome coronavirus helicase (MERS-CoV nsp13) | | Descriptor: | ORF1ab, SULFATE ION, ZINC ION | | Authors: | Hao, W, Wojdyla, J.A, Zhao, R, Han, R, Das, R, Zlatev, I, Manoharan, M, Wang, M, Cui, S. | | Deposit date: | 2017-01-03 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Middle East respiratory syndrome coronavirus helicase
PLoS Pathog., 13, 2017
|
|
3HXG
 
 | | Crystal structure of Schistsome eIF4E complexed with m7GpppA and 4E-BP | | Descriptor: | Eukaryotic Translation Initiation Factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | Authors: | Liu, W, Zhao, R, Jones, D.N.M, Davis, R.E. | | Deposit date: | 2009-06-20 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into parasite EIF4E binding specificity for m7G and m2,2,7G mRNA cap.
J.Biol.Chem., 284, 2009
|
|
3HXI
 
 | | Crystal structure of Schistosome eIF4E complexed with m7GpppG and 4E-BP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Eukaryotic Translation Initiation 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Liu, W, Zhao, R, Jones, D.N.M, Davis, R.E. | | Deposit date: | 2009-06-20 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into parasite EIF4E binding specificity for m7G and m2,2,7G mRNA cap.
J.Biol.Chem., 284, 2009
|
|
3HZH
 
 | | Crystal structure of the CheX-CheY-BeF3-Mg+2 complex from Borrelia burgdorferi | | Descriptor: | Chemotaxis operon protein (CheX), Chemotaxis response regulator (CheY-3), MAGNESIUM ION | | Authors: | Pazy, Y, Silversmith, R.E, Guarinari, M, Zhao, R. | | Deposit date: | 2009-06-23 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HIB
 
 | |
3CGZ
 
 | |
3CGY
 
 | | Crystal Structure of Salmonella Sensor Kinase PhoQ catalytic domain in complex with radicicol | | Descriptor: | RADICICOL, Virulence sensor histidine kinase phoQ | | Authors: | Guarnieri, M.T, Zhang, L, Shen, J, Zhao, R. | | Deposit date: | 2008-03-06 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Hsp90 inhibitor radicicol interacts with the ATP-binding pocket of bacterial sensor kinase PhoQ.
J.Mol.Biol., 379, 2008
|
|
3E66
 
 | | Crystal structure of the beta-finger domain of yeast Prp8 | | Descriptor: | PRP8 | | Authors: | Yang, K, Zhang, L, Xu, T, Heroux, A, Zhao, R. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3QI3
 
 | | Crystal structure of PDE9A(Q453E) in complex with inhibitor BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
3QI4
 
 | | Crystal structure of PDE9A(Q453E) in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
2QIC
 
 | | Crystal Structure of the ING1 PHD Finger in complex with a Histone H3K4ME3 peptide | | Descriptor: | H3K4ME3 PEPTIDE, Inhibitor of growth protein 1, ZINC ION | | Authors: | Pena, P.V, Champagne, K, Zhao, R, Kutateladze, T.G. | | Deposit date: | 2007-07-03 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor.
J.Mol.Biol., 380, 2008
|
|
1AYN
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Oliveira, M.A, Zhao, R, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human rhinovirus 16.
Structure, 1, 1993
|
|
1I7T
 
 | | CRYSTAL STRUCTURE OF CLASS I MHC A2 IN COMPLEX WITH PEPTIDE P1049-5V | | Descriptor: | 9 RESIDUE PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Busslep, J, Zhao, R, Loftus, D, Appella, E, Collins, E.J. | | Deposit date: | 2001-03-10 | | Release date: | 2001-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | T cell activity correlates with oligomeric peptide-major histocompatibility complex binding on T cell surface
J.Biol.Chem., 276, 2001
|
|
1I7R
 
 | | CRYSTAL STRUCTURE OF CLASS I MHC A2 IN COMPLEX WITH PEPTIDE P1058 | | Descriptor: | 9 RESIDUE PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Busslep, J, Zhao, R, Loftus, D, Appella, E, Collins, E.J. | | Deposit date: | 2001-03-10 | | Release date: | 2001-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | T cell activity correlates with oligomeric peptide/MHC binding on T cell surface
J.Biol.Chem., 276, 2001
|
|
