1MIH
 
 | | A ROLE FOR CHEY GLU 89 IN CHEZ-MEDIATED DEPHOSPHORYLATION OF THE E. COLI CHEMOTAXIS RESPONSE REGULATOR CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, MANGANESE (II) ION, ... | | Authors: | Silversmith, R.E, Guanga, G.P, Betts, L, Chu, C, Zhao, R, Bourret, R.B. | | Deposit date: | 2002-08-23 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89.
J.Bacteriol., 185, 2003
|
|
3HZH
 
 | | Crystal structure of the CheX-CheY-BeF3-Mg+2 complex from Borrelia burgdorferi | | Descriptor: | Chemotaxis operon protein (CheX), Chemotaxis response regulator (CheY-3), MAGNESIUM ION | | Authors: | Pazy, Y, Silversmith, R.E, Guarinari, M, Zhao, R. | | Deposit date: | 2009-06-23 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5D2C
 
 | | Reaction of phosphorylated CheY with imidazole 1 of 3 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Page, S, Silversmith, R.E, Bourret, R.B, Collins, E.J. | | Deposit date: | 2015-08-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Imidazole as a Small Molecule Analogue in Two-Component Signal Transduction.
Biochemistry, 54, 2015
|
|
5DGC
 
 | | Reaction of phosphorylated CheY with imidazole 2 of 3 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, Chemotaxis protein CheY, ... | | Authors: | Page, S, Silversmith, R.E, Bourret, R.B, Collins, E.J. | | Deposit date: | 2015-08-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Imidazole as a Small Molecule Analogue in Two-Component Signal Transduction.
Biochemistry, 54, 2015
|
|
5DKF
 
 | | Reaction of phosphorylated CheY with imidazole 3 of 3 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Page, S, Silversmith, R.E, Bourret, R.B, Collins, E.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Imidazole as a Small Molecule Analogue in Two-Component Signal Transduction.
Biochemistry, 54, 2015
|
|
1KMI
 
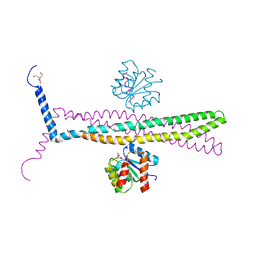 | | CRYSTAL STRUCTURE OF AN E.COLI CHEMOTAXIS PROTEIN, CHEZ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BICINE, Chemotaxis protein cheY, ... | | Authors: | Zhao, R, Collins, E.J, Bourret, R.B, Silversmith, R.E. | | Deposit date: | 2001-12-16 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ.
Nat.Struct.Biol., 9, 2002
|
|
1RLM
 
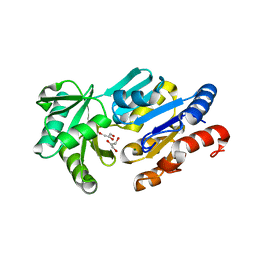 | | Crystal Structure of ybiV from Escherichia coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLO
 
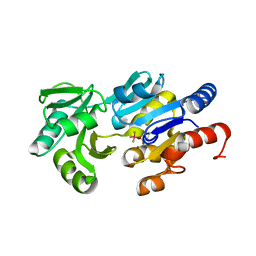 | | Phospho-aspartyl Intermediate Analogue of ybiV from E. coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLT
 
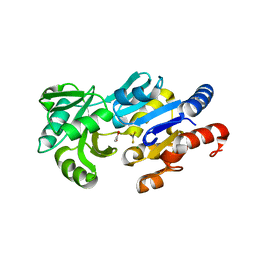 | | Transition State Analogue of ybiV from E. coli K12 | | Descriptor: | ACETATE ION, ALUMINUM FLUORIDE, GLYCEROL, ... | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
3OO0
 
 | | Structure of apo CheY A113P | | Descriptor: | AMMONIUM ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Guanga, G.P, Miller, P.J, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2010-08-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring the effect of an allosteric site on conformational coupling in CheY
To be Published
|
|
3RVK
 
 | | Structure of the CheY-Mn2+ Complex with substitutions at 59 and 89: N59D E89Q | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVP
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89K | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVJ
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89Q | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVO
 
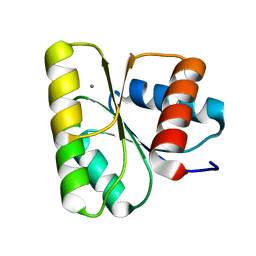 | | Structure of CheY-Mn2+ Complex with substitutions at 59 and 89: N59D E89Y | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVM
 
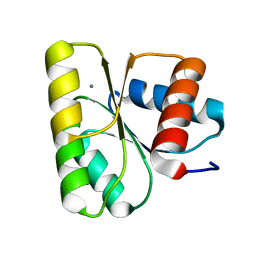 | | Structure of the CheY-Mn2+ Complex with substitutions at 59 and 89: N59D and E89R | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVN
 
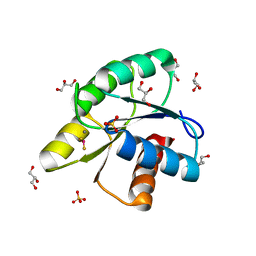 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89Y | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVS
 
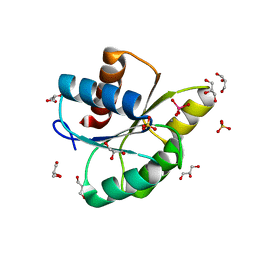 | | Structure of the CheYN59D/E89R Tungstate complex | | Descriptor: | Chemotaxis protein CheY, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVR
 
 | | Structure of the CheYN59D/E89R Molybdate complex | | Descriptor: | Chemotaxis protein CheY, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVL
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89R | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVQ
 
 | | Structure of the CheY-Mn2+ Complex with substitutions at 59 and 89: N59D E89K | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
