8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
8GXH
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
7VUX
 
 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
8GXI
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14c | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of SARS-CoV-2 main protease in complex with 14c
To Be Published
|
|
8GX3
 
 | | The crystal structure of human Calpain-1 protease core in complex with 14c | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14c
To Be Published
|
|
8GX2
 
 | | The crystal structure of human CtsL in complex with 14c | | Descriptor: | DIMETHYL SULFOXIDE, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human CtsL in complex with 14a
To Be Published
|
|
7W33
 
 | | The crystal structure of human CtsL in complex with 14a | | Descriptor: | N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2R,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
7W34
 
 | | The crystal structure of human CtsL in complex with 14b | | Descriptor: | N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
8TM5
 
 | | Human mixed 13S proteasome assembly intermediate | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Zhang, H, Zhao, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human 20S proteasome biogenesis.
Nat Commun, 15, 2024
|
|
8TM4
 
 | | Human pre 13S proteasome assembly intermediate | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Zhang, H, Zhao, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human 20S proteasome biogenesis.
Nat Commun, 15, 2024
|
|
8TM3
 
 | | Human proteasome alpha ring assembly intermediate | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome assembly chaperone 3, ... | | Authors: | Zhang, H, Zhao, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human 20S proteasome biogenesis.
Nat Commun, 15, 2024
|
|
8TM6
 
 | | Human premature 20S proteasome assembly intermediate | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Zhang, H, Zhao, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of human 20S proteasome biogenesis.
Nat Commun, 15, 2024
|
|
5GAR
 
 | | Thermus thermophilus V/A-ATPase, conformation 1 | | Descriptor: | Archaeal/vacuolar-type H+-ATPase subunit I, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Schep, D.G, Zhao, J, Rubinstein, J.L. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GAS
 
 | | Thermus thermophilus V/A-ATPase, conformation 2 | | Descriptor: | Archaeal/vacuolar-type H+-ATPase subunit I, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Schep, D.G, Zhao, J, Rubinstein, J.L. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5H3J
 
 | |
5YS1
 
 | | Crystal structure of Multicopper Oxidase CueO G304K mutant | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | Deposit date: | 2017-11-12 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
5YS5
 
 | | Crystal structure of Multicopper Oxidase CueO G304K mutant with seven copper ions | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | Deposit date: | 2017-11-13 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
5I1M
 
 | | Yeast V-ATPase average of densities, a subunit segment | | Descriptor: | V-type proton ATPase subunit a, vacuolar isoform | | Authors: | Schep, D.G, Zhao, J, Rubinstein, J.L. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7W7O
 
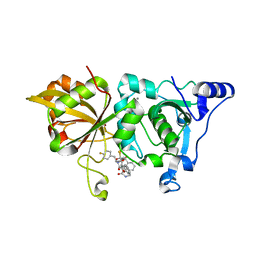 | | The crystal structure of human Calpain-1 protease core in complex with 14a | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, HYDROSULFURIC ACID, ... | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2021-12-06 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14a
To Be Published
|
|
5HXD
 
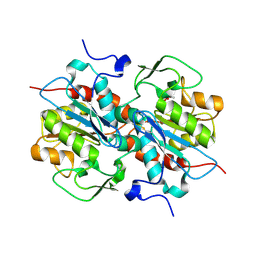 | | Crystal structure of murein-tripeptide amidase MpaA from Escherichia coli O157 | | Descriptor: | CACODYLATE ION, Protein MpaA, ZINC ION | | Authors: | Ma, Y, Bai, G, Zhang, X, Zhao, J, Yuan, Z, Kang, X, Li, Z, Mu, S, Liu, X. | | Deposit date: | 2016-01-30 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Murein-Tripeptide Amidase MpaA from Escherichia coli O157 at 2.6 angstrom Resolution
Protein Pept.Lett., 24, 2017
|
|
2N4H
 
 | |
2N3X
 
 | |
8IVQ
 
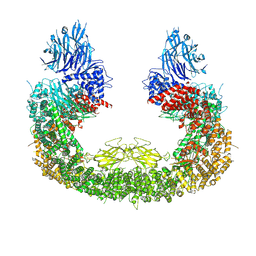 | | Cryo-EM structure of mouse BIRC6, Global map | | Descriptor: | Isoform 2 of Baculoviral IAP repeat-containing protein 6 | | Authors: | Liu, S, Jiang, T, Bu, F, Zhao, J, Wang, G, Li, N, Gao, N, Qiu, X. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms underlying the BIRC6-mediated regulation of apoptosis and autophagy.
Nat Commun, 15, 2024
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
